Abstract
COVID-19 has been challenging to stop because the virus was new to humans and keeps changing in ways that make it spread faster and cause disease in many people, which is called an epidemic. Health authorities and doctors must hurry to decide which protection measures, like closing borders or developing vaccines, will work to fight each kind of harmful microbe depending on how dangerous and widespread it is. Fortunately, a new scientific technique called whole genome sequencing can quickly spot and track pathogens—microbes that cause infectious diseases. Whole genome sequencing works well for both disease-causing viruses and bacteria. This technique can help scientists discover new epidemics and reveal how diseases are spreading, aiding health authorities in their quest to stop epidemics much faster. As diseases around the world are being tracked using this method, we stand a better chance of limiting future epidemics like COVID-19.
What Causes Infectious Diseases?
Many infectious diseases (diseases that can spread between people or from animals to people) are caused by bacteria and viruses. These are extremely small microbes: to see bacteria, we need to use a microscope with 1,000-times magnification. To see viruses, we need a more powerful type of microscope called an electron microscope, which can magnify things 10,000–100,000 times. There are millions of species of viruses and bacteria that inhabit the Earth, but only a few of these microbes cause disease in humans, animals, or plants. Disease-causing microbes are also called pathogens. Pathogens have caused diseases in humans for thousands of years. Some infectious diseases are mild, like the common cold, but others are deadly. At any time, new pathogens can suddenly appear and cause diseases that humans have never been exposed to before—like when an animal virus “crosses over” to infect humans, for example. This is what happened in 2019, when the coronavirus named SARS-CoV-2 started infecting humans, causing COVID-19.
Epidemics and Pandemics—When Infectious Diseases Spread
Pathogens can spread between people in various ways. For instance, pathogens like SARS-CoV-2 spread from an infected person to others nearby through coughing, sneezing, or even just speaking—actions that send tiny, virus-containing droplets flying through the air. Other pathogens, like bacteria called Salmonella that can cause diarrhea, can spread when humans eat foods contaminated with these bacteria.
When a disease spreads quickly throughout a group of people, it is called an epidemic. An epidemic can occur, for example, when a bunch of people eat the same contaminated food product and all become infected with Salmonella, or when many people in one region get COVID-19. A pandemic is an epidemic that spreads worldwide. Epidemics and pandemics happen when a pathogen finds a lot of people who are susceptible to infection because they have never been exposed to that pathogen before.
How are Epidemics Spotted?
Government public health services help to prevent human diseases by detecting and stopping epidemics. To do so, these organizations have created public health surveillance systems that help them to count the number of cases of certain diseases happening in various locations and among specific groups of people, like children and adults. For example, public health surveillance systems track COVID-19 and Salmonella infections in people around the world. These disease surveillance systems are connected with other surveillance systems that test for pathogens in the environment, such as the presence of Salmonella in food and water.
Identifying the pathogen causing a new epidemic is urgent because controlling the pathogen’s spread depends on knowing what it is, as well as how and where it is transmitted. Pathogen identification is also needed to develop tests, treatments, and vaccines. Public health surveillance systems are designed to detect epidemics quickly and stop them as soon as possible. For that reason, public health surveillance systems must continually receive test results from the labs that doctors use to identify pathogens in samples (blood, urine, throat or nose swabs) from sick patients. When they detect an emerging epidemic, public health officials can then inform the public and other health professionals about their findings. You may remember hearing these official reports on TV or seeing them on the internet during the height of the COVID-19 pandemic.
Infectious Diseases Change Over Time
One thing that can make epidemics and pandemics hard to control is that infectious diseases change over time. While humans take about 20 years to have children, bacteria and viruses reproduce much more quickly. It takes as little as 8 h for viruses to multiply, and bacteria are even faster—some can double in number as often as every 20 min! Every time a pathogen reproduces, it makes a copy of its entire genome for its descendants. Each time a copy is made, small copying errors called mutations can happen in the genome. These mutations can change the characteristics of the offspring. Thus, mutations can create new pathogen variants (also called strains). Variants may differ from the parent and from other strains in certain characteristics, which might change the ways they infect people or respond to tests, treatments, or vaccines. Pathogen variants happen all the time, but only some mutations help pathogens to spread and create more offspring. This is a process of evolution called natural selection.
The rapid evolution of pathogens explains why variants of SARS-CoV-2 are still popping up today, causing more cases of COVID-19. Some SARS-CoV-2 variants have caused huge epidemic waves around the world. These variants differ in how easily they spread among people and how severe their disease symptoms are. Potentially dangerous variants are called variants of public health concern, and they are assigned a Greek letter, like Delta or Omicron, to help scientists track them. When experts identify the emergence of a new, dangerous variant, urgent public health measures (like wearing face masks or closing borders) may have to be taken to limit the damage these variants cause. Scientists and drug companies must also use information from public health surveillance systems to quickly make new vaccines and treatments to protect people against emerging variants. It is like an arms race between the virus and health professionals.
Genomic Surveillance: A New, Fast Way to Identify Pathogens and Detect Epidemics
From what you have learned so far, it is probably clear to you that quick detection of new pathogens or emerging variants is extremely important for protecting public health. The faster these dangerous organisms can be detected and characterized, the better!
Tracking pathogens by looking at their genomes is called genomic surveillance [1]. In the past, it took a lot of work and time for scientists to test for pathogens in medical samples. Some testing was done by growing bacteria or viruses in the lab or by copying pathogen genes many times, with a technique called polymerase chain reaction (PCR). These time-consuming techniques allowed researchers to study only one or a few types of pathogen in a sample (and only if that pathogen had been previously identified), or to look at only a few mutations at a time. However, in recent years, scientists and engineers have invented a new technique that can identify all pathogens and new variants much more quickly. This technique is called whole genome sequencing (WGS). WGS uses very accurate instruments to “read” the entire genome of a single virus or bacterium from a sample in one go. This is much faster and easier than the old way, giving results in a matter of hours instead of days. It is run by a robot, and the information that is generated has a lot more details. For instance, the genome sequence provided by WGS can tell scientists if the pathogen can be treated with certain antibiotics, or which vaccine can protect people against it. WGS can also provide information that allows the development of PCR tests or other rapid tests. The process of genomic surveillance is illustrated in Figure 1.
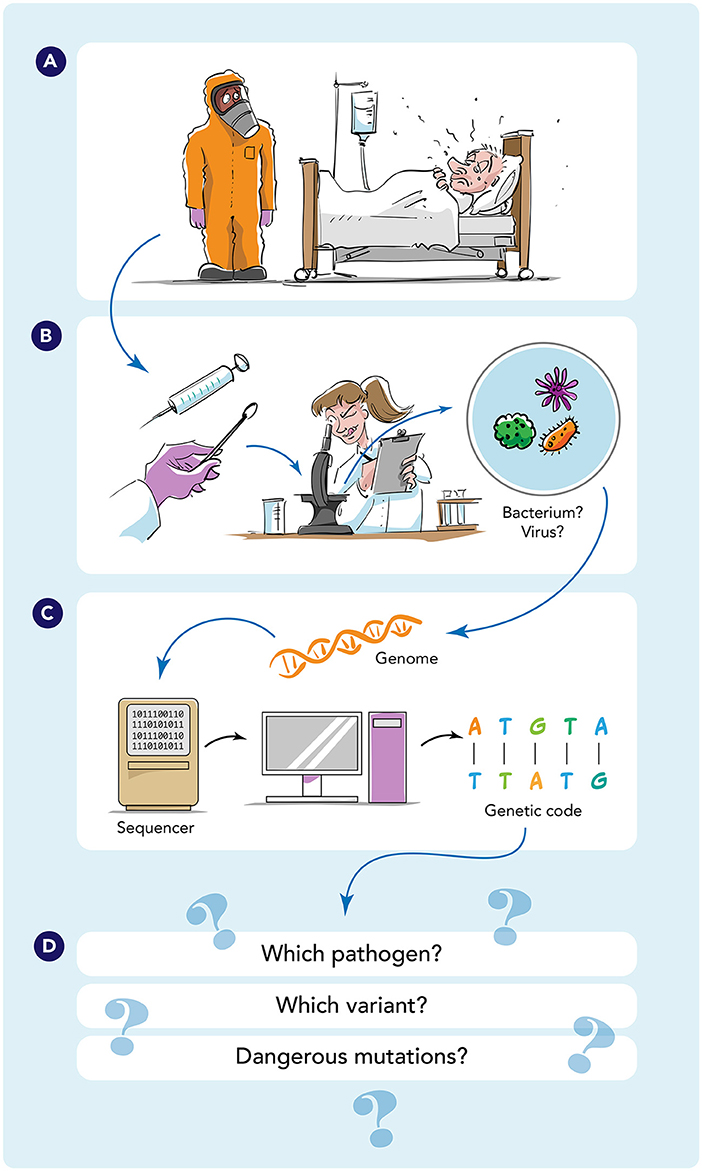
- Figure 1 - Process of genomic surveillance.
- (A) When patients have an infectious disease, medical professionals take samples of blood, saliva, or cells. (B) These samples are analyzed in a lab to determine whether the patient is infected with a virus or bacteria. (C) To identify the pathogen (e.g., SARS-CoV-2 or Salmonella), scientists can use WGS to read the pathogen’s entire genome. (D) WGS helps scientists identify the pathogen, determine whether it is a variant (and which one), and know whether the pathogen contains a mutation that might make it resistant to medicines or vaccines, for example.
Using WGS, scientists can identify any viral or bacterial pathogen by the sequence of its genome. They do that using a combination of biology, computer science, and math to understand data and recognize patterns, a method called bioinformatics. Using bioinformatics to compare the unknown sequence to a database of known genome sequences allows them to detect the unique “signature sequence” for any species of virus or bacteria. WGS can also detect any mutations in a pathogen’s genome, which makes it the best way to quickly identify new pathogen variants and track variants of public health concern [1]. What makes this technique even more powerful is that it can be tweaked to allow scientists to read all the genomes from all the microbes present in medical samples (blood or saliva from sick people, for example) or from the environment (like ocean water or city wastewater samples), which is called metagenomics [2].
Studying Wastewater to Predict Epidemics
Another benefit of genomic surveillance is that it helps public health surveillance systems all over the world to spot new epidemics very early [1]. This can be done by looking at data on who was sick, where, and when, and combining that information with data on the genome sequence of the pathogen from sick people. When several people are found to be infected with the same pathogen (identical genome) over a short time, this means they may have infected each other, or possibly been infected from the same source. Such information can give experts a warning sign of an emerging epidemic, which they can then further investigate—kind of like detectives investigating a crime, who find fingerprints that help them to identify the suspect. This helps to stop a pathogen in its tracks, before it causes much damage.
One interesting way to do this is to perform metagenomics on wastewater—water that has been used in homes, schools, and businesses [3]. But how does this work? When people have COVID-19, they release SARS-CoV-2 genomes into the sewer system in their poop and pee. In populated areas that send their wastewater to treatment facilities, scientists can take samples of the wastewater for metagenomic analysis. Using metagenomics and bioinformatics, scientists can detect viruses, identify variants and any mutations, and measure how many genomes they find in the wastewater. These findings tell them when new variants appear or when a variant increases in frequency in the nearby community. Tracking variants of SARS-CoV-2 in wastewater has been effective in showing public health officials when a variant of SARS-CoV-2 was starting a new epidemic in a certain area [3].
One Health: Genomic Surveillance for a Healthier World
Genomic surveillance is helping experts to prevent epidemics of infectious diseases like COVID-19 and Salmonella in many countries, stopping diseases in their tracks much faster than was possible before [4]. Genomic surveillance works best when many ill people can get medical tests to identify the pathogens making them sick, and when patient samples and medical data are quickly transported to public health authorities. Genomic surveillance of animals, as well as testing for dangerous microbes in food and water, can provide even more data to help prevent epidemics. This is part of a concept called One Health, which recognizes that human health, animal health, and the health of the environment are all connected—so all three aspects must be addressed to track and prevent epidemics (Figure 2) [5]. Unfortunately, many countries still do not have enough testing across these three areas to make genomic surveillance as accurate as it could be.
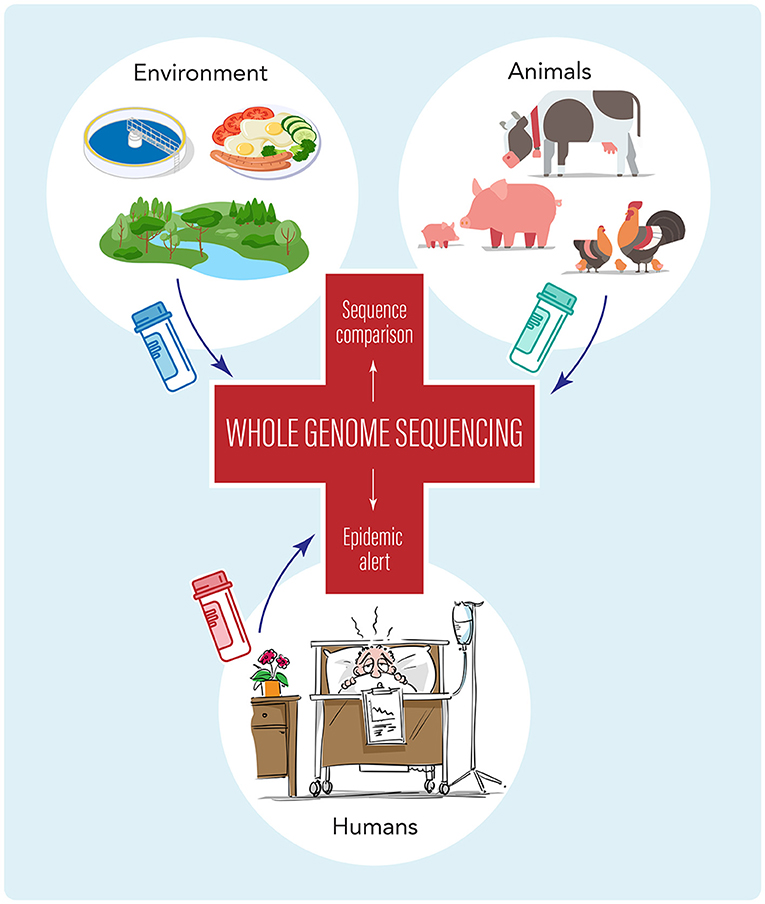
- Figure 2 - The health of humans, animals, and the environment are all connected.
- The concept of One Health tells us that the best way to address and prevent epidemics involves genomic surveillance of sick people, animals (including birds, pets, and farm animals), and aspects of the environment (like food, water, and wastewater). This is done by comparing the genome sequences of microbial pathogens to find matches between samples, which can warn of pathogens moving between these three areas, indicating possible new disease epidemics.
Since genomic surveillance helps public health authorities quickly take the actions needed to stop epidemics, it is important to make these techniques available in every country in the world. Genomic surveillance is especially important in countries where infectious diseases are more frequent and dangerous, such as countries where many people live in poverty and do not have access to a strong healthcare system. Pathogens do not respect borders between countries, so collaboration between countries in detecting and stopping epidemics with up-to-date genomic surveillance technology benefits everyone, everywhere in the world. Hopefully, wise use of this amazing technology will help prevent another pandemic disaster like COVID-19 from happening in the future [5].
Glossary
Pathogen: ↑ A microbe that can cause disease.
Epidemic: ↑ A rapid increase of new cases of infectious disease in a population, driven by uncontrolled spread of the pathogen that causes the disease.
Pandemic: ↑ An epidemic that spreads worldwide.
Public Health Surveillance Systems: ↑ Organizations that collect and analyze data on human diseases, to provide information about actions necessary to protect public health.
Genome: ↑ The full set of genetic information of an organism.
Variants: ↑ Different versions of a pathogen, like a virus or bacteria, that have small genetic changes. These changes can affect how the pathogen spreads or causes disease.
Genomic Surveillance: ↑ Studying pathogen genomes along with information on who is sick, where, and when, to detect and trace epidemics.
Whole Genome Sequencing: ↑ A relatively new way to “read” the entire genome of an organism, like a virus or bacterium.
Metagenomics: ↑ “Reading” the genomes from many microbes at once, to identify those present in samples from humans (like medical samples) or from the environment (like water).
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Edited by Susan Debad Ph.D., graduate of the University of Massachusetts Graduate School of Biomedical Sciences (USA) and scientific writer/editor at SJD Consulting, LLC. We would like to thank the coauthors of the original manuscript: Catherine Ludden, Guido Werner, Vitali Sintchenko, Pikka Jokelainen, and Margaret Ip.
Original Source Article
↑ Struelens, M. J., Ludden, C., Werner, G., Sintchenko, V., Jokelainen, P., and Ip, M. 2024. Real-time genomic surveillance for enhanced control of infectious diseases and antimicrobial resistance. Front. Sci. 2:1298248. doi: 10.3389/fsci.2024.1298248
References
[1] ↑ Gardy, J. L., and Loman, N. J. 2018. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 19:9–20. doi: 10.1038/nrg.2017.88
[2] ↑ Ko, K. K. K., Chng, K. R., and Nagarajan, N. 2022. Metagenomics-enabled microbial surveillance. Nat. Microbiol. 7:486–96. doi: 10.1038/s41564-022-01089-w
[3] ↑ Levy, J. I., Andersen, K. G., Knight, R., and Karthikeyan, S. 2023. Wastewater surveillance for public health. Science 379:26–7. doi: 10.1126/science.ade2503
[4] ↑ Chattaway, M. A., Dallman, T. J., Larkin, L., Nair, S., McCormick, J., Mikhail, A., et al. 2019. The transformation of reference microbiology methods and surveillance for salmonella with the use of whole genome sequencing in England and Wales. Front Public Health. 7:317. doi: 10.3389/fpubh.2019.00317
[5] ↑ Aarestrup, F. M., Bonten, M., and Koopmans, M. 2021. Pandemics- one health preparedness for the next. Lancet Reg. Health Eur. 9:100210. doi: 10.1016/j.lanepe.2021.100210