Abstract
Salmonella is an important type of bacteria capable of infecting humans and animals. These bacteria were discovered over 100 years ago, and thousands of types have now been identified. Salmonella can be found in many sources, and its pathway to causing disease in humans is a complex web. Salmonella can live in farm, wild, and domesticated animals; the environment; and water sources—all of which can contaminate food items and, in consequence, humans. The most frequent illness in humans is called salmonellosis, which is characterized by diarrhea, stomach cramps, fever, nausea, vomiting, and headache. Although most people have mild symptoms, a few cases may evolve into a more severe illness that may need medicines called antibiotics to treat. In this article, you will learn about the importance of Salmonella, as well as the challenges it poses for the future and promising scientific advances that will improve our knowledge of these bacteria.
What is Salmonella?
Salmonella are important bacteria that can cause illness in humans and can also infect wild and domesticated animals [1]. They belong to a large family of bacteria called Enterobacteriaceae, which is very important in the medical field due to its ability to infect humans. Under the microscope, Salmonella strains have a rod-like shape (Figure 1). They can survive and grow in the presence or absence of oxygen, and they use sugars as their energy source.
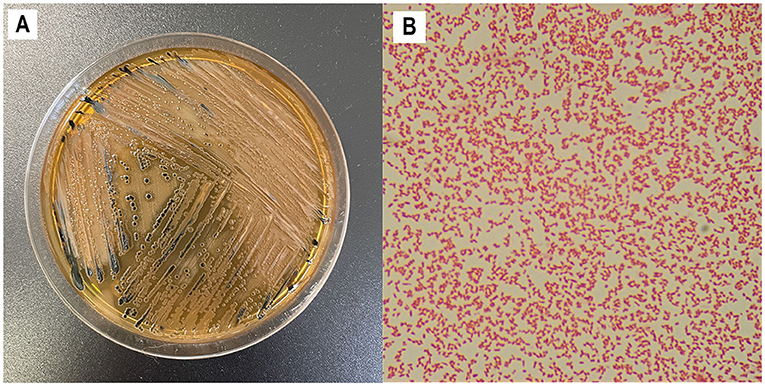
- Figure 1 - Growing and examining Salmonella.
- (A) Salmonella forms growth dots, called colonies, when it is grown on Jello-like substances, called agar plates. In this figure, you can see how Salmonella grows in Salmonella-Shigella agar. (B) Dyes can be used in a method, called Gram staining, to give Salmonella a pink/red color and allow it to be seen under a microscope. If you look closely, you can see the rod-like shape of each Salmonella cell.
Salmonella were first described back in 1884 by two scientists, Daniel Elmer Salmon and Theobald Smith, while they were studying the effects of a severe infection in the intestines of pigs. But it was not until 1900 that another scientist, Joseph Lignières, named these bacteria “Salmonella,” as a way to honor the studies of Daniel Salmon.
The Complicated Classification of Salmonella
Salmonella have a really extensive and complex classification system [1]. They are divided into two species, called S. bongori and S. enterica. Both are commonly found in cold-blooded animals (such as reptiles) and in the environment. However, one of the six subspecies of S. enterica (which is also called S. enterica) is particularly important, as it can infect several warm-blooded animals, such as birds and mammals, including humans (Figure 2).
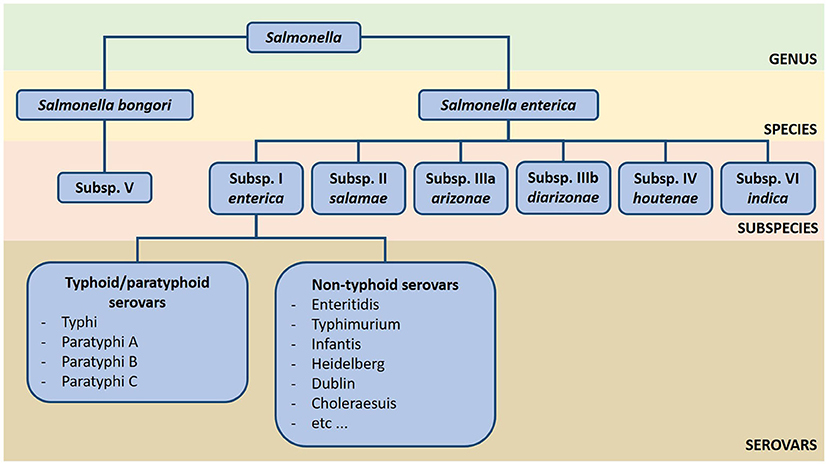
- Figure 2
- An overview of the extensive classification system of Salmonella. The genus Salmonella has two species, S. bongori and S. enterica, that can also be divided in seven distinct subspecies. S. enterica subspecies enterica is commonly found in warm-blooded animals (such as mammals and birds), while the other subspecies are more present in the environment and cold-blooded animals (such as reptiles). Subspecies enterica currently has more than 1.5 thousand serovars identified, which can be classified as typhoid/paratyphoid and non-typhoid serovars according to the two types of diseases they cause in humans: typhoid/paratyphoid fever and salmonellosis, respectively.
Scientists classify Salmonella using a method called serotyping. In the laboratory, we get a small scoop of the growing Salmonella bacteria and mix them with small drops of a substance called serum. These serums come from animals, and their molecules can bind to different structures present in the bacterial cell outer membrane. If the Salmonella react to specific combinations of serums, the bacteria are classified into a group called a serovar (Figure 2). Using this method, scientists have identified and named more than 2.6 thousand serovars of Salmonella so far [1].
We can also categorize Salmonella serovars according to what illnesses they cause in humans: typhoid/paratyphoid Salmonella cause diseases called typhoid or paratyphoid fever, while non-typhoid Salmonella cause salmonellosis (Figure 2) [1]. The rest of this article will focus on the importance of non-typhoid Salmonella.
How Animals and Foods Are Linked to Transmission of Salmonella
If you knew about Salmonella before reading this article, you most likely related it to eating contaminated raw meat or eggs. However, the dangerous relationship of Salmonella and food is bigger than we imagine, and it begins long before we are eating or cooking our food (Figure 3) [2].
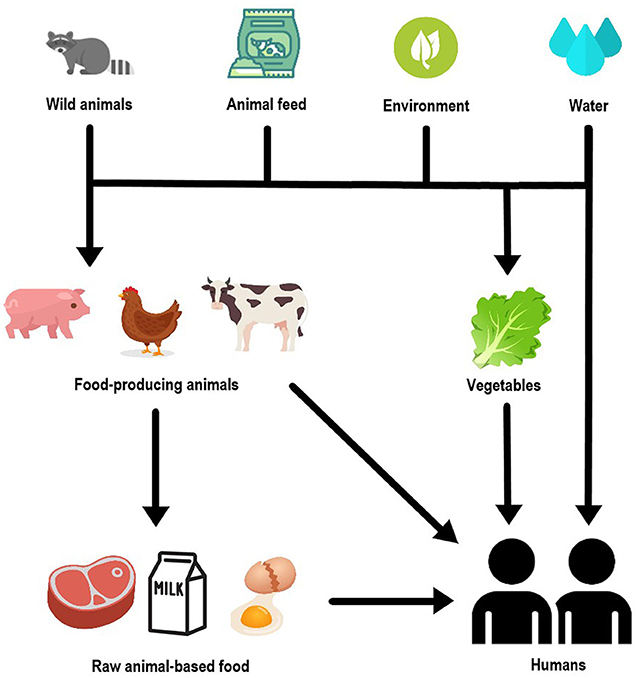
- Figure 3 - Non-typhoid Salmonella can move among humans, animals, and foods in various ways.
- Salmonella is mainly transmitted to humans by contaminated food, especially those of animal origin, but also vegetables and water. Food-producing animals can become contaminated with Salmonella through diverse sources, such as contact with wild animals, the environment, and contaminated water or feed given to these animals. Finally, it is also worth to mention that humans can get Salmonella after having contact with contaminated animals.
Chickens are the main source of non-typhoid Salmonella among farm animals. However, pigs, cows, and turkeys have also been important animal sources of non-typhoid Salmonella. Their meat and related food products (such as ham, sausages, milk, and cheese) can be potential sources of the bacteria [2].
But how do these farm animals get contaminated with non-typhoid Salmonella? There are a number of ways. One of them is through the environment—but not only the natural environment. Farm environments and industry settings where meat and food items are produced can be sources of contamination, too. Some studies also show that animal feed (including the foods we give to pets) can be contaminated with non-typhoid Salmonella because some types of feed are produced with animal meat. In addition, contact with wild animals can also be a potential route by which farm animals can become contaminated. Wild animals and even insects have also been found to carry non-typhoid Salmonella [2].
We usually eat vegetables fresh, but if they are not carefully washed, vegetables can also be sources of non-typhoid Salmonella. Vegetables usually get contaminated water contaminated with bacteria is used to water crops, or they can get contaminated from the water used to clean them, if adequate cleaners are not used [2]. Farm animals (and people) can also get non-typhoid Salmonella by drinking contaminated water, and people can also get it through handling infected farm animals.
To prevent people from getting sick due to Salmonella in food, simple measures can be used. The U.S. Centers for Disease Control and Prevention (CDC) suggests that food be cooked to a minimum temperature of 145–165°F (~62–74°C), depending of the type. Eating raw or undercooked (“runny”) eggs should be avoided. Perishable foods should be stored in the fridge, at temperatures under 40°F (~4°C). It is also important to use adequate cleaning methods for food and cooking instruments, and to practice good hand washing habits. You can find these and other useful information about Salmonella and food at the CDC website, at https://www.cdc.gov/foodsafety/communication/salmonella-food.html.
Salmonellosis: What it is and How to Treat it
Salmonellosis is the main human illness caused by non-typhoid Salmonella. In the United States alone, the CDC estimates that 1.35 million infections, 26,500 hospitalizations, and 420 deaths occur every year. The World Health Organization (WHO) considers non-typhoid Salmonella to be one of the four main bacteria that cause diarrhea in humans.
The first symptoms usually appear 12–36 h after eating contaminated food, but this period may vary from as short as 6 h up to 6 days. The most classic symptom of salmonellosis is diarrhea, which is a watery, loose stool (poop), usually accompanied by stomach cramps and fever. In more severe cases, blood can also be present in the diarrhea. Nausea, vomiting, and headaches can also happen sometimes.
The symptoms are usually fairly mild and they generally last 4–7 days. Some people have cases with such mild symptoms that the infection even goes unnoticed. However, these bacteria can be found in the feces of infected individuals for several weeks after symptoms end. Most patients naturally recover by simply drinking a lot of fluids, which prevents dehydration caused by diarrhea.
Severe cases of dehydration can be especially concerning for children under 2 years old and for elderly patients. These cases can result in symptoms such as long-lasting diarrhea and vomiting, high fever (above 102°F/38.5°C), bloody stool, and dehydration indicators such as very little urine, dry mouth, and dizziness. In these severe cases, people should get medical treatment. They might need to receive fluids directly into their veins to become rehydrated.
A very small number of patients develop severe cases of what is called invasive salmonellosis. In these situations, the bacterial infection is not limited to the gut. Instead, Salmonella may infect the blood, the membranes surrounding the brain and spinal cord, and the bones and joints. Severe illnesses are more common in people with weakened immune systems, such as children under the age of 2 years, elderly people, people living with HIV, cancer, or autoimmune diseases, or people who take certain medications. In these cases, hydration alone is not enough to treat salmonellosis. Instead, antibiotics are needed to ensure a patient’s survival.
You can find these and more information about the burden, disease, symptoms and treatment of Salmonella infections at the CDC (https://www.cdc.gov/salmonella/general/index.html) and WHO [https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal)] websites.
The Future of Salmonella Studies
The future of Salmonella studies may bring great advances but serious challenges [3, 4]. For example, as reported by the WHO and CDC, a growing number of bacteria now have antibiotic resistance, including Salmonella. Imagine this: a patient infected with Salmonella develops a serious illness, so doctors start treatment with antibiotics. However, the patient’s condition does not improve, and suddenly their life is at risk. Sad and worrying, right? That is why antibiotic resistance is so concerning and must be prevented. New ways to understand and prevent the spread of bacteria causing disease in humans, as well as hard work to discover new antibiotics, are goals for the future [3].
There is reason to hope that we will have ways to respond to dangerous Salmonella infections. Over the years, the study of bacterial DNA has taught scientists a huge amount about Salmonella, and even faster techniques are now available. Imagine how many questions might be quickly answerable when the entire DNA sequences of certain bacteria are in a computer file. Scientists could see whether a sample of bacteria is related to others already identified in other countries or time periods. They could investigate whether the bacteria have any genes that might make them resistant to antibiotic, or genes that might indicate the severity of the illness they will cause [4]. Such information will be very helpful to scientists and doctors.
Final Remarks
Now you know why non-typhoid Salmonella are such important bacteria. Thousands of serovars capable of infecting humans and farm animals have already been identified. You also learned about the impact of Salmonella in food, and its spider web of transmission routes that includes humans, several types of food items, farm and wild animals, and various environments. Salmonellosis in humans is an illness with global impact, and it can vary from mild to severe. However, treatment of salmonellosis may be at risk due to increasing antibiotic resistance. Finally, we also briefly examined how new DNA technologies are helping scientist to understand Salmonella.
We hope that this article provided you with useful information about Salmonella that you can use in your daily life. Our even greater hope is that this information awakens your curiosity and inspires you to learn more about these bacteria. The challenges of the future are already happening, and we will need more curious and smart minds to help in our battle against Salmonella!
Glossary
Serotyping: ↑ A laboratory technique that uses serums that bind to surface structures of the bacterial cell to identify them. Salmonella are classified in serovars if they react to specific serum combinations.
Serum: ↑ Liquid obtained from the blood of animals, which is rich in antibodies (molecules that protect against infectious and non-infectious agents).
Serovar: ↑ A group of bacteria classified as belonging to a specific type through the serotyping method, these types are called “serotype” or “serovar”.
Salmonellosis: ↑ Disease caused by non-typhoid Salmonella. The main symptoms are diarrhea, stomach cramps, fever, and sometimes nausea, vomiting, and headache.
Contamination: ↑ Undesirable elements or impurities that infect or corrupt materials, organisms, environments, etc.
Antibiotics: ↑ Drugs developed to kill or prevent the multiplication of bacteria in the human body.
Antibiotic Resistance: ↑ The ability of bacteria to survive in the presence of antibiotics. Bacteria usually become resistant through changes in their DNA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by a research grant from the São Paulo Research Foundation (Proc. 22/07013-0) under the supervision of JF. During the course of this work, FV was supported by a PhD student scholarship (Proc. 141017/2021-0) and JF by a Productive fellowship (Proc. 304399/2018-3 and 304803/2021-9) from the National Council for Scientific and Technological Development. This study was financed by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil—Finance Code 001.
References
[1] ↑ Jajere, S. M. 2019. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World 12:504–21. doi: 10.14202/vetworld.2019.504-521
[2] ↑ Chanamé Pinedo, L., Mughini-Gras, L., Franz, E., Hald, T., and Pires, S. M. 2022. Sources and trends of human salmonellosis in Europe, 2015-2019: an analysis of outbreak data. Int. J. Food Microbiol. 379:109850. doi: 10.1016/j.ijfoodmicro.2022.109850
[3] ↑ McDermott, P. F., Tyson, G. H., Kabera, C., Chen, Y., Li, C., Folster, J. P., et al. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother. 60:5515–0. doi: 10.1128/AAC.01030-16
[4] ↑ Pornsukarom, S., van Vliet, A. H. M., and Thakur, S. 2018. Whole genome sequencing analysis of multiple Salmonella serovars provides insights into phylogenetic relatedness, antimicrobial resistance, and virulence markers across humans, food animals and agriculture environmental sources. BMC Genom. 19:801. doi: 10.1186/s12864-018-5137-4
