Abstract
Life has been evolving in the oceans much longer than it has on land, resulting in highly diverse ocean organisms—particularly microbes like bacteria and archaea. Ocean microbes perform crucial functions that influence the health of the ocean and ultimately impact Earth’s climate. To understand the diversity and functions of marine organisms, scientists have used a powerful technique called metagenomics to study the DNA of all the organisms present in an ocean-water sample at once. In our research, we combined results from multiple ocean metagenomic studies, taken from various locations and depth zones across the world’s oceans, to produce a global ocean genome composed of 317.5 million groups of similar genes—approximately half of which could be categorized by type of organism and function. This unprecedented amount of data has much to teach us about varied ocean habitats and can help scientists answer many questions about ocean organisms and their functions.
The Ocean—The Cradle of Life
The ocean is the world’s largest habitat. Oceans cover 71% of our planet’s surface and contain an absolutely enormous volume of water-1.335 billion km3, about 97% of all the water on Earth! The ocean is also the cradle of life—Earth’s first life forms appeared in this watery world about 3.9 billion years ago.
Many kinds of organisms live in the ocean, from plants to animals to bacteria to viruses. While you are probably most familiar with larger marine animals like whales, dolphins, and octopi, the ocean actually contains a large array of animal life. Earth’s first life forms originated in the ocean and only later evolved to conquer land. The long evolutionary history of ocean-based life is illustrated by the fact that, of the 34 major known animal phyla, there is only one that exists exclusively on land (that’s Onychophora, a kind of worm that has legs and a texture like velvet) [1]. It should not be surprising, given how long life has been evolving there, that the ocean contains a tremendous biodiversity—much of which remains unexplored.
All animals—those in the ocean and those on land—are classified as eukaryotes, which means that each of their cells has a distinct nucleus and other membrane-bound functional units called organelles. However, by far the most abundant organisms in the ocean are the prokaryotes, ocean microbes that are invisible to the naked eye. Prokaryotes, which are single-celled organisms without nuclei or specialized organelles, include two types of organisms: bacteria, which you have probably heard of, and archaea, which you might be less familiar with. While bacteria and archaea have many things in common, one important feature that sets these two groups apart is that the cell walls of many bacteria contain a substance called peptidoglycan, which the cell walls of archaea completely lack.
While we know that there are more than two million species of bacteria in the global ocean, we still do not know much about them or the other types of ocean microbes. These organisms are extremely difficult to study because more than 99% of them have not been grown in the lab!
Why Are Ocean Microbes Important?
Individual species of bacteria and archaea are fascinatingly varied. Importantly, they have many unique ways of obtaining energy. Metabolism is the word for the energy-producing chemical reactions inside most cells. These reactions use “food” sources to produce the energy that cells need to grow, reproduce, and generally stay alive. Some bacteria and archaea use sunlight to produce energy, while others break down or transform compounds containing carbon, nitrogen, or sulfur—elements that can affect ocean chemistry and that also play important roles in Earth’s changing climate [2, 3].
Additionally, ocean microbes can be sources of molecules that play key roles in biotechnology. For example, a protein called Taq polymerase was isolated from bacteria growing in super-hot ocean water near hydrothermal vents, which are like deep-sea geysers. This protein is now an important part of a common laboratory test called PCR, which is used for many scientific and medical studies including the detection of the SARS-CoV-2 virus that causes COVID-19.
Just from these examples, you can see that, despite their tiny size, ocean microbes—particularly bacteria and archaea—are critically important ocean-dwellers. What is even more intriguing is that many ocean organisms and their associated functions have not yet been discovered. How can we get a picture of all the organisms that live in the ocean—from eukaryotes to the smallest and hardest-to-study prokaryotes? It would be great, would it not, if we could simply take samples of water or sediment from the ocean and quickly analyze them in the lab to find out what kinds of organisms are living there and which functions they perform? Well, thanks to a powerful new technique called metagenomics, researchers can do this!
Studying Genes to Understand Organisms
Every living organism contains DNA, which holds the instructions—genes—that code for the structure and function of that organism. Collectively, all of an organism’s genes are called its genome.
One of the main ways that scientists study the genomes of organisms (a field called genomics) is by sequencing their DNA. Sequencing is a laboratory technique that allows scientists to “read” the DNA code. For many years, DNA sequencing was an error-prone, time-consuming process and scientists could only sequence short stretches of DNA in each experiment—it took an awfully long time to read the genome of an organism. For example, sequencing of the entire human genome, called the Human Genome Project, took 13 years of work by many scientists spread across 20 institutions worldwide [4].
However, in the past decades, some amazing technical advances in the field of DNA sequencing have not only made whole-genome sequencing faster, easier, and cheaper, but have enabled the field of metagenomics to explode. “Meta-” means “higher” or “beyond,” so, metagenomics is “beyond” the study of one organism’s genome—it is the study of the pooled genetic information from all the organisms contained in a sample from the environment, like water or soil. Using metagenomics techniques, scientists can sequence all the DNA in a sample of ocean water to figure out the global ocean genome—all the genes from all the organisms in that sample. This information can tell scientists which types of organisms are present and what kinds of ecologically important functions they are performing in ocean habitats [5]!
The Power of Metagenomics: Who Lives in the Ocean and What Are They Doing?
The first metagenomic study of the ocean was performed by the Sorcerer II Global Ocean Sampling Expedition, back in 2003–2004, which analyzed a marine plankton community. Over the past 10 years, other expeditions have occurred, including the TARA Ocean Expedition, which collected 243 samples from 68 sampling sites between 2009 and 2013, mostly from the upper ocean. Metagenomics analysis of these samples identified 33.3 million genes! (In contrast, the human genome contains approximately 30,000 genes—see here for more info.) Additional studies, including the Malaspina Expedition, the Ocean Sampling Day Program [6], and the Hawaii Ocean Time Series, have expanded on these results, sampling other oceans, specific marine habitats, and water from various depths.
It is important to recognize that the ocean environment is not the same from the surface to the ocean floor—we can divide it into four unique zones, as shown in Figure 1A. In our research, we combined the vast amount of metagenome data from 2,112 ocean samples collected from multiple past studies (Figures 1B, C), to create a full “catalog” of the global ocean genome, called KMAP Global Ocean Gene Catalog 1.0. This catalog contains an amazing amount of information-317.5 million gene clusters (collections of similar genes grouped together according to function), approximately half of which are annotated, meaning they have been classified according to the type of organism they belong to and the functions of the proteins they code for.
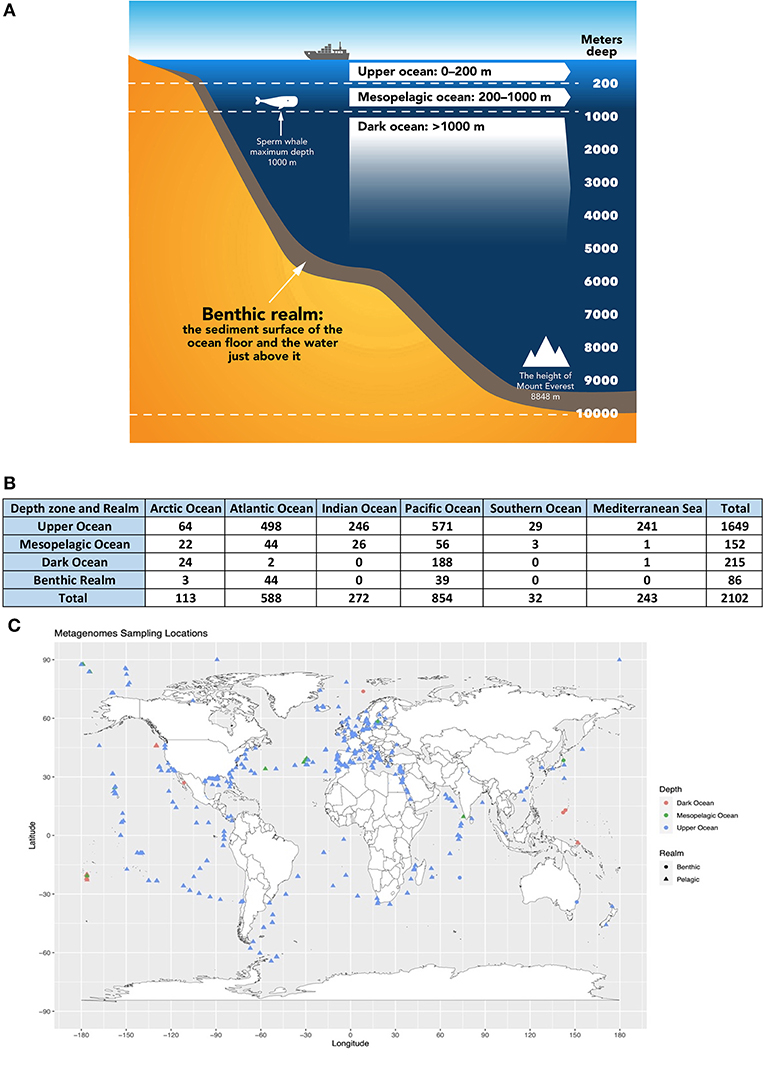
- Figure 1 - Samples compiled for our study.
- (A) Ocean zones and depths. (B) Locations and depth zones of the samples used for our research. You can see that certain oceans, like the Southern Ocean, and certain depth zones, like the dark ocean and the benthic realm, are underrepresented in these samples. (C) Locations from which the metagenomes used in our research were obtained across the benthic and pelagic realms. Depth zones are shown by the colors of the circles.
Bacteria: The Ocean’s Major Players
Focusing on the annotated gene clusters—those that we can link to a particular type of organism and for which we have functional information—we found that, across all ocean samples, 78.34% of the annotated genes were from bacteria, followed by eukaryotes (12.16%), archaea (6.05%), and viruses (3.44%) (Figure 2A). While genes from bacteria were dominant in every depth zone, genes from these four major types of ocean organisms were not evenly distributed throughout all zones—the benthic realm and the various depth zones of the pelagic realm contained differing proportions of each organism type (Figure 2B). This is not surprising because conditions across depth zones vary—particularly in the amount of light that is available—creating a variety of unique ecological niches in which specific types of organisms can thrive.
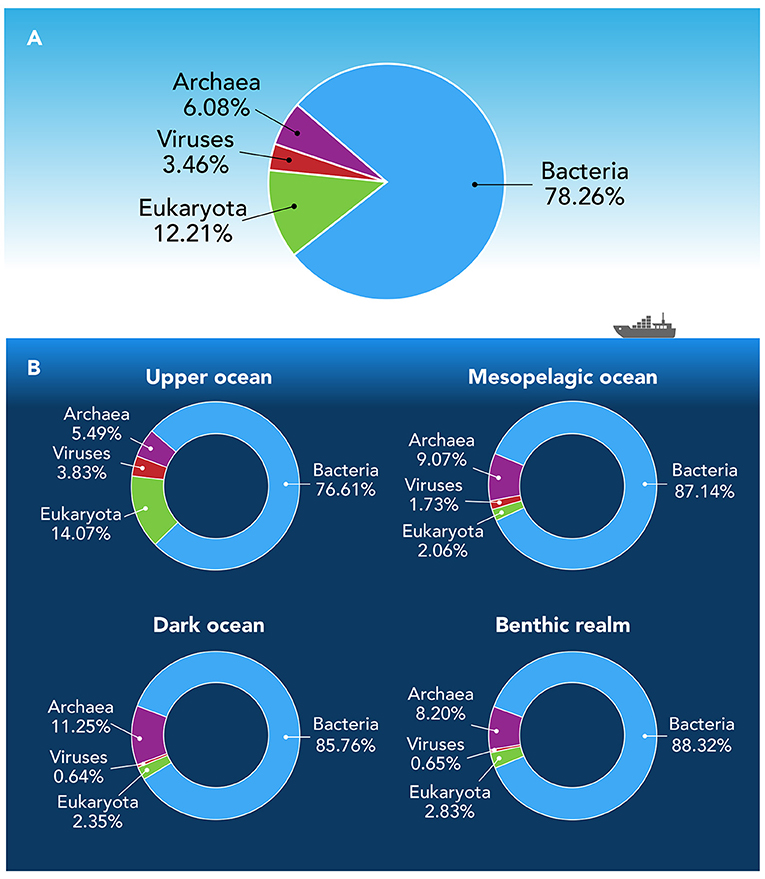
- Figure 2 - (A) Overall abundance of genes from the four major types of organisms across all ocean samples.
- (B) The abundance of genes from each type of organism differs between depth zones of the pelagic realm (upper, mesopelagic, and dark ocean) and the benthic realm. This tells us that each region of the ocean forms a unique habitat with specific growth conditions, in which certain organisms can thrive.
Microbe Metabolism Can Influence Earth’s Climate
Microbial metabolism keeps the oceans healthy by controlling the flow of nutrients and energy. As we mentioned earlier, some microbial metabolic processes influence the cycling of elements—like carbon—that can influence Earth’s climate. Given the importance of microbial metabolism, we took an in-depth look at the metabolism-related genes in our catalog. In samples from the benthic realm, 41.3% of the annotated gene clusters were involved in metabolic processes, compared to just 25% of the gene clusters from the pelagic communities. This tells us that there is a lot going on in this commonly neglected and understudied region of the ocean!
When we sorted the annotated metabolism-related genes found across all ocean samples by the substances being metabolized, we found that that 50% of those genes took part in some form of carbon-related metabolism (Figure 3). Carbon-containing molecules, like carbon dioxide (CO2) and methane (CH4) are greenhouse gases that contribute to global warming. Some bacteria and algae can use photosynthesis to turn CO2 into carbohydrates, with help from sunlight, thus removing CO2 from the atmosphere. But photosynthesis is not the only pathway that ocean organisms use to metabolize carbon! Methane-metabolizing pathways, for example, do not require light and can function in the deep ocean and benthic realm, where light does not penetrate. Some of these pathways have been discovered quite recently and scientists still have much to learn about them.
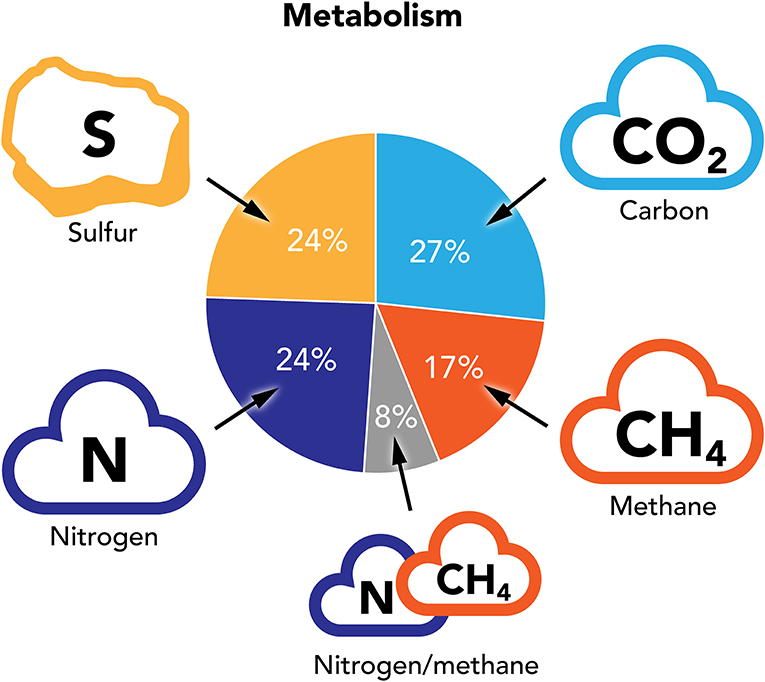
- Figure 3 - Across all ocean samples, more than 50% of the annotated metabolism genes identified in our metagenomic analysis were related to some form of carbon metabolism (CO2 or CH4).
- Genes involved in the metabolism of nitrogen and sulfur were also present.
Metagenomics: The Future of Ocean Research?
The KMAP Ocean Gene Catalog 1.0 consists of approximately 163 million annotated gene clusters, providing us with an extraordinary amount of information about the types of organisms living at various depths of the ocean and the functions they perform. The global ocean genome is much more than just a simple catalog of the many organisms living there and their functions—it can deepen our understanding of the ocean and provide us with a “baseline” from which to evaluate the effects of human disturbances that can upset the ocean’s delicate balance, including pollution, overfishing, and ocean mining.
While sequencing and studying the global ocean genome has been occurring for some time, two factors have limited the size of those studies: technology and cost. Recent breakthroughs in sequencing technologies have resulted in systems that are faster, more accurate, and portable [7]. Portability is especially important to marine scientists because portable systems can be used onboard oceangoing research vessels. These advances in sequencing technology have also dramatically reduced its cost. In the past, scientists had to ask smaller questions and carefully choose which sequencing experiments to perform because this research was extremely expensive. Today, modern, inexpensive sequencing technology allows relatively simple collection of genome data on an unprecedented scale. These data can help scientists answer questions about the vast array of ecosystem services that ocean organisms provide to keep our entire planet—and us—healthy.
Do you have questions about ocean organisms and their functions? Maybe future metagenomic studies of the global ocean genome will provide answers!
Glossary
Eukaryote: ↑ An organism whose cells contain a nucleus and other membrane-bound functional units called organelles. Animals, plants, and fungi are examples of eukaryotes.
Prokaryote: ↑ A single-celled organism that does not have a distinct nucleus or organelles. Bacteria and archaea are prokaryotes.
Peptidoglycan: ↑ A substance forming the cell walls of many bacteria. Peptidoglycans are not present in the cell walls of archaea.
Metabolism: ↑ Chemical reactions within cells that change “food” into energy.
Genome: ↑ The collection of all the genes of an organism.
Sequencing: ↑ A laboratory technique that allows scientists to “read” the DNA code, either small segments or the entire genome of an organism.
Metagenomics: ↑ The study of the genomes of all the organisms in a sample, such as a sample of ocean water or those on the human skin.
Annotated: ↑ Classified according to the type of organism the genes belong to and the functions of the proteins they code for.
Greenhouse Gases: ↑ Gases in the Earth’s atmosphere that trap the sun’s heat, preventing it from escaping back into space.
Global Warming: ↑ The rising of Earth’s average temperature caused by greenhouse gases in the atmosphere.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by King Abdullah University of Science and Technology through competitive center funding provided to the Computational Bioscience Research Center. Co-written by Susan Debad Ph.D., graduate of the University of Massachusetts Graduate School of Biomedical Sciences (USA) and scientific writer/editor at SJD Consulting, LLC.
Original Source Article
↑Laiolo, E., Alam, I., Uludag, M., Jamil, T., Agusti, S., Gojobori, T., et. al. 2024. Metagenomic probing toward an atlas of the taxonomic and metabolic foundations of the global ocean genome. Front. Sci. 1:1038696. doi: 10.3389/fsci.2023.1038696
References
[1] ↑ Jaume D., and Duarte C. M. 2006. General Aspects Concerning Marine and Terrestrial Biodiversity. The Exploration of Marine Biodiversity–Scientific and Technological Challenges. Bilbao: Fundación BBVA.
[2] ↑ Venter, J. C., Remington, K., Heidelberg, J. F., Halpern, A. L., Rusch, D., Eisen, J. A., et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. doi: 10.1126/science.1093857
[3] ↑ Rusch, D. B., Halpern, A. L., Sutton, G., Heidelberg, K. B., Williamson, S., Yooseph, S., et al. 2007. The sorcerer II global ocean sampling expedition: Northwest Atlantic through eastern tropical pacific. PLoS Biol. 5:e77. doi: 10.1371/journal.pbio.0050077
[4] ↑ Wikipedia. 2022. Human Genome Project. Available online at: https://en.wikipedia.org/wiki/Human_Genome_Project (accessed January 18, 2023).
[5] ↑ Escobar-Zepeda, A., Godoy-Lozano, E. E., Raggi, L., Segovia, L., Merino, E., Gutiérrez-Rios, R. M., et al. 2018. Analysis of sequencing strategies and tools for taxonomic annotation: defining standards for progressive metagenomics. Sci. Rep. 8:12034. doi: 10.1038/s41598-018-30515-5
[6] ↑ Kopf, A., Bicak, M., Kottmann, R., Schnetzer, J., Kostadinov, I., Lehmann, K., et al. 2015. The ocean sampling day consortium. GigaSci. 4:27. doi: 10.1186/s13742-015-0066-5
[7] ↑ Tyler, A. D., Mataseje, L., Urfano, C. J., Schmidt, L., Antonation, K. S., Mulvey, M. R., et al. 2018. Evaluation of oxford nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Sci. Rep. 8:10931. doi: 10.1038/s41598-018-29334-5
