Abstract
The ocean is the home of many amazing living things such as fishes, shrimps, jellyfish, and seagrasses. You might have seen some of these organisms in the ocean or a beautiful aquarium. But did you know that there is another kind of marine life that dramatically outnumbers all other ocean organisms, but they are so small that you cannot even see them? They are microbes! Microbes are important for the ocean because some of them turn waste produced by other organisms back into food, and others can even influence Earth’s climate. Scientists want to learn more about ocean microbes. However, it is super hard to grow these microbes in an aquarium or a lab because we need to know exactly what they like to eat and what their favorite conditions are. What can we do? We use a cool technique called metagenomics, which can help us figure out which microbes are found in the ocean by putting together genetic puzzles!
Why Should We Care About Ocean Microbes?
Earth is different from other planets because it has many amazing living things. More than 70% of Earth’s surface is covered by seawater, and many organisms live in the ocean. Fishes, shrimps, or seagrasses might quickly come to mind when you think about marine life. However, there is one group of organisms that dramatically outnumbers any of the others, but they are so tiny that we cannot see them with our eyes. These very small life forms called microbes. One ml of seawater near the ocean surface has around one million microbes! These microbes include some “bad” ones that make people sick, but most ocean microbes help keep the ocean healthy and are important to sustain life on Earth.
Here are examples of those environmentally relevant marine microbes. Carbon dioxide is a greenhouse gas, and too much carbon dioxide in the atmosphere warms Earth faster than living things can handle. Cyanobacteria are a group of marine phytoplankton (see this Frontiers for Young Minds article), which are tiny ocean organisms similar to plants. With the help of sunlight, phytoplankton take in carbon dioxide to grow, and in the process they release oxygen. Without the actions of phytoplankton, Earth would warm up even faster. Other types of microbes, like ocean bacteria that take in oxygen, are also important. They can break down dead organisms and the wastes organisms produce, and turn these materials back into nutrients that phytoplankton can use to grow [1]. Small shrimps then feed on phytoplankton and the shrimps are part of the diets of certain fishes, forming the food web. Without ocean microbes, the entire ocean food web would collapse.
Some Marine Microbes Live Without Oxygen
Some marine microbes live in unique ocean regions where oxygen cannot be detected—areas where living things that do depend on oxygen cannot live (Figure 1). These microbes use other chemicals instead of oxygen to “breath”, such as different forms of nitrogen (see this Frontiers for Young Minds article) [2], and they can change nitrogen compounds that are essential food for other ocean organisms into gasses, which then escape from the ocean. Thus, microbes in oxygen-free seawater control the rate at which marine life grows and multiplies throughout the ocean. Additionally, one type of released nitrogen gasses, called nitrous oxide, is a greenhouse gas with a warming power around 300 times stronger than carbon dioxide. So, you can see that microbes living in oxygen-free regions of the ocean play several very important roles in keeping our planet healthy and in balance.
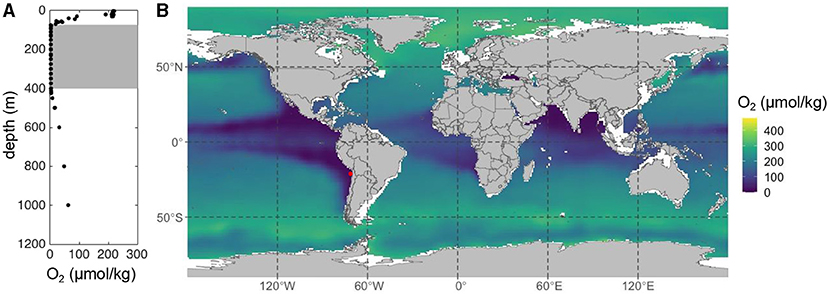
- Figure 1 - Oxygen concentrations in the ocean.
- (A) In the Eastern Tropical South Pacific [red dot in (B), where we sampled [3]], oxygen concentration changes with depth. Gray area indicates oxygen-free seawater. (B) Oxygen concentrations at 200 m depth in the world’s oceans (oxygen data from World Ocean Atlas). The dark purple areas are those special oceanic regions with low or no oxygen.
Marine Microbes Are Hard to Grow in the Lab
Most microbes are single cells, so you would think they would be very easy to grow and take care of compared to complex organisms with many cells, such as puppies. In some cases, this is true—some bacteria can double themselves within only a few hours at room temperature, if they are given some simple nutrients. However, growing marine microbes is usually not easy. One reason is that most of the ocean has low concentrations of nutrients. Thus, the most abundant and successful microbes in the ocean are usually those that are good at taking up very tiny amounts of nutrients, but these microbes grow very slowly. Even though ocean nutrient concentrations are low, there are many kinds of nutrients in the ocean. Marine microbes often need a variety of nutrients to grow. Due to the vast diversity but low concentration of ocean nutrients, it is hard to figure out which nutrients ocean microbes need to grow and thrive.
Microbes from seawater without oxygen are exceptionally hard to grow. On top of the difficulties we already mentioned, scientists must ensure that no oxygen is around while they are growing these microbes. That is not easy since nearly 20% of the air is oxygen. Even the systems scientists can purchase to seal bottles to create oxygen-free environments are saturated with oxygen. Consequently, growing microbes in the lab can only help us understand a small proportion of ocean microbes, especially in the oxygen-free parts of the ocean. How can we learn about the diverse microbes that have yet to be successfully grown in the lab?
Putting Together Genetic Puzzles to Study Hard-to-Grow Microbes
Thanks to the development of metagenomics, we can now get to know microbes by examining their genetic material, which tells us what kinds of functions they may be performing—without the need to grow them (Figure 2). Metagenomics is the exploration of the genetic information of all microbes collected from a certain environment (e.g., ocean, land, rivers, lakes, and inside plants and animals—including humans). To study ocean microbes using metagenomics, scientists collect marine microbes by running liters of seawater through filters with tiny holes, which let seawater run through but trap the microbes (Figure 2). Then, the genetic material of the microbes is separated from other cell parts and is sequenced using laboratory equipment that reveals the genetic code of each microbe (the latter process is called sequencing; Figure 2).
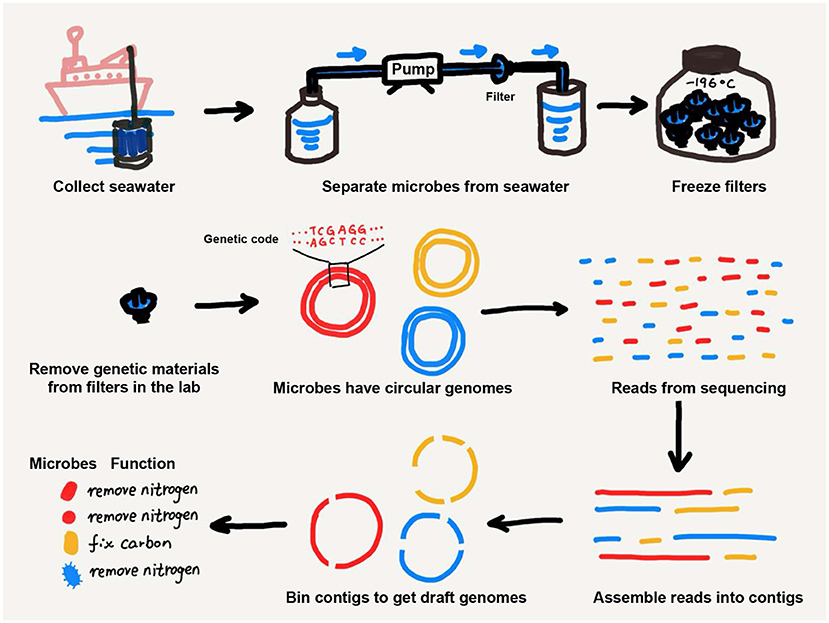
- Figure 2 - Metagenomics involves sampling microbes from the ocean and putting together genetic puzzles to understand which microbes are present in the ocean and what they can do.
The genetic material of each microbe is mainly stored in a circular genome. However, during sample collection and sequencing, circular genomes break into short pieces (we call them reads). The reads in our samples belonging to different microbes get mixed up together. To understand the genomes of the different kinds of microbes in our samples, we must link these short pieces (i.e., reads) into longer pieces of genetic material (a process known as assembly) and then sort these longer pieces according to which microbes they belong to (a process known as binning). This entire process is like putting together a puzzle with millions of pieces (Figure 2).
Putting Together the Genetic Puzzles
The first step is called assembly. Assembly involves connecting short reads into longer pieces (we call these longer pieces contigs) based on their overlapping parts. For example, if the end of a read is the same as the beginning of another read, we know they were originally linked and belong to the same genome. However, assembly does not give us all the genomes of marine microbes because some parts of each genome are usually lost during sampling and sequencing, meaning there are gaps in the genomes where we have no reads.
The next step is binning, which is putting all the contigs that we cannot link any further into genomes. Like pieces from the same jigsaw puzzle, contigs originally belonging to the same microbial genome share some similarities. The genetic material of living organisms is made up of a combination of four different molecules (called nucleotides), abbreviated A, T, C, and G. Each organism has its own preference for using certain nucleotides more than others, or for putting certain nucleotides next to each other. These preferences can help scientists find contigs from the same genome. Other information can also be used to group contigs into genomes. After binning, we have microbial genomes that still have some gaps compared to the actual genomes of the microbes, and we call them draft genomes or metagenome-assembled genomes (we will use “draft genomes” hereinafter) to differentiate them from the full genomes without gaps.
What Did We Learn About Ocean Microbes From Playing Genetic Puzzles?
Draft genomes created by putting together genetic puzzles can provide valuable information about microbes that cannot be grown in the lab. In a recent study, we recovered draft genomes from seawater in the Eastern Tropical South Pacific where there is very little or no oxygen (Figure 1) [3]. By comparing these draft genomes with those from the global ocean [4], we found that there are many microbes living in low- or no-oxygen seawater. More than half of the microbes we found in our samples had never been discovered before (Figure 3). These draft genomes suggest that many of these microbes use nitrogen when oxygen is not available, and some nitrogen might be turned into gases that leave the ocean and enter the atmosphere (Figure 3).
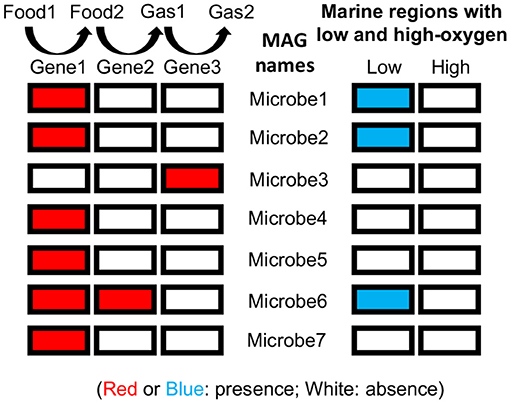
- Figure 3 - Nitrogen-breathing genes and representation of MAGs in the global ocean.
- In the ocean, nitrogen is present in forms that serve as food for living things. The process of converting “food” nitrogen to gas forms has several steps. Our research showed that specific microbes can perform specific steps in the process, controlled by different genes. Presence (red) and absence (white) of genes involved in this process are shown for each draft genome. On the right, you can see whether these draft genomes were found in other ocean regions with low or high oxygen where other scientists collected metagenomic data [modified from Figure 2 in Sun and Ward [3]].
As mentioned earlier, nitrogen is an essential nutrient for all living things. So, understanding how much nitrogen is turned into gases and lost from the ocean is important for scientists to estimate the amount of living things that the ocean can support. One main nitrogen-loss process has multiple steps. Previous knowledge of this process was largely based on microbes that can grow in the lab, and most of those lab microbes can perform all the steps. If most microbes associated with this process in low-oxygen seawater can also perform the entire process, they could cause loss of nitrogen from the ocean. By examining draft genomes from low-oxygen seawater, however, we found that most microbes associated with the nitrogen-loss process could only perform some of the steps. This means that many of these microbes do not cause nitrogen loss from the ocean. The new microbes that we discovered using metagenomics tell us that we must revisit the way we estimate nitrogen loss from the ocean.
Glossary
Microbes: ↑ Tiny organisms that are too small to be seen with our eyes. Most microbes are single-cell organisms such as bacteria and archaea.
Phytoplankton: ↑ Tiny organisms live in water and act like plants. They use energy from sunlight to turn carbon dioxide and water into oxygen, and they grow during this process.
Metagenomics: ↑ The exploration of all genetic information from all the organisms collected from a certain environment. Metagenomics does not target just one type of genetic information or one type of organisms; instead, it examines a mix of genetic information from various organisms.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
XS was supported by the Simons Foundation postdoctoral fellowship in Marine Microbial Ecology.
Original Source Article
↑Sun, X., and Ward, B. B. 2021. Novel metagenome-assembled genomes involved in the nitrogen cycle from a Pacific oxygen minimum zone. ISME Commun. 1:26. doi: 10.1038/s43705-021-00030-2
References
[1] ↑ Pomeroy, L. R., leB. Williams, P. J., Azam, F., and Hobbie, J. E. 2007. The microbial loop. Oceanography 20:28–33. doi: 10.5670/oceanog.2007.45
[2] ↑ Ward, B. B. 2013. Oceans. How nitrogen is lost. Science 341:352–3. doi: 10.1126/science.1240314
[3] ↑ Sun, X., and Ward, B. B. 2021. Novel metagenome-assembled genomes involved in the nitrogen cycle from a Pacific oxygen minimum zone. ISME Commun. 1:26. doi: 10.1038/s43705-021-00030-2
[4] ↑ Tully, B. J., Graham, E. D., and Heidelberg, J. F. 2018. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci. Data 5:170203. doi: 10.1038/sdata.2017.203
