Abstract
Viruses are unique biological systems. They are parasites that use the cells of other organisms, called hosts, to multiply, often causing disease to the host. One of the most interesting features of viruses is that some of them contain RNA as their genetic material—all other known organisms use DNA. In the beginning of my career I worked on RNA viruses, trying to understand their basic behaviors and processes. When I focused on RNA viruses that are known to cause cancer, I discovered that they can make DNA from their RNA genomes, in a process called reverse transcription. That was a great discovery that changed the prevailing way of thinking and had profound implications in the fields of biology, medicine, and biotechnology. In this article, I will tell you about viruses, walk you through the discovery of reverse transcription, and describe some major implications of our findings in terms of improving or even saving many human lives.
Professor David Baltimore won the Nobel Prize in Physiology or Medicine in 1975, jointly with Prof. Renato Dulbecco and Prof. Howard M. Temin, for their discoveries concerning the interaction between tumor viruses and the genetic material of the cell.
The Fascinating World of Viruses
Viruses are tiny particles that infect cells and sometimes cause disease. Viruses exist in uncountable numbers, and every living animal and even every bacterium has its own set of viruses. More than a million different viruses have been identified! You are probably familiar with some viruses, such as the influenza viruses that cause flu; SARS-CoV-2, which causes COVID-19; or varicella-zoster virus, which causes chickenpox. Maybe you have heard of the human immunodeficiency virus (HIV), which causes AIDS, or the Ebola virus. Viruses vary dramatically in their structures (Figure 1), in how many species they can infect (whether they infect only a particular species or several species), and how dangerous they are to the organisms they infect, which are called the hosts.
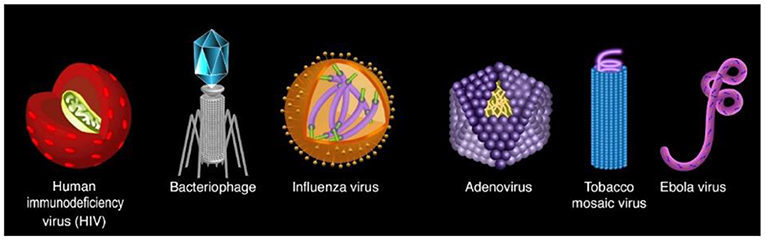
- Figure 1 - The variety of viruses.
- Viruses come in many different shapes. Every animal and bacterium has its own set of viruses (Image adapted from here).
Viruses multiply by penetrating a host cell and inserting their genetic material, which the host then treats as its own. This way, viruses “enslave” the host cell, tricking it into producing many copies of the virus. Viruses multiply extremely quickly—in as little as 20–30 min in bacteria and a few hours in mammals, including humans. They spread by moving from one host to another. For example, some viruses can move between humans through sneezing or when someone touches a surface that was previously touched by an infected person. We can defend ourselves from some viruses using vaccination, and viruses that infect only humans are easier to handle.
As you might know, DNA is the molecule that carries the genetic information in living systems. DNA has a relative called RNA, which serves as a moveable copy of the information in DNA and enables the production of proteins (To learn more about DNA, RNA, and proteins, see this Nobel Collection article about protein degradation and this Nobel Collection article about RNA splicing.). All organisms in the animal kingdom have DNA as their genetic material. Viruses are unique in that some of them, called RNA viruses, use RNA as their genetic material (It is believed that viruses are remnants of an earlier world, in which RNA was used as the genetic information. To read more about this early RNA world, read this article.). DNA and RNA viruses have various ways of using their genetic material to make a specific type of RNA called messenger RNA (mRNA) [1], which is used to make proteins. I created a classification system for viruses, called the Baltimore Classification, which classifies viruses based on the way they make mRNA. Within this classification, there are two types of DNA viruses that use only DNA to make mRNA (groups I, II), three types of RNA viruses that use only RNA to make mRNA (groups III, IV, V), and two groups of viruses that use both DNA and RNA to make mRNA (groups VI, VII) [1, 2].
Studying RNA Viruses
When I started my scientific career in the 1960s, I wanted to work on the fundamental chemistry of life. It seemed to me that viruses offer the best opportunity to do that because they are the simplest organisms in the world. We could study viruses and understand their operation in detail—all the way down to the molecular level.
In the 1960s, we knew very little about how RNA viruses replicate. At first, I worked on a polio-like RNA virus that grows in mice (group IV according to the Baltimore Classification, quite similar to SARS-CoV-2), trying to understand how this virus multiplies and how it affects the life of its host. I deciphered the replication mechanism of this polio-like virus and then extended the findings to other RNA viruses. In this process, I discovered several important proteins called enzymes that make DNA and RNA [3, 4].
Around 1970, I became interested in whether there were other, undiscovered ways that RNA viruses replicate themselves. I was particularly interested in RNA viruses that cause cancer, called RNA tumor viruses. When I started to study these viruses, I had no idea that a great surprise was awaiting me.
Reverse Transcription
In the 1960s, a colleague of mine named Howard Temin suggested that RNA tumor viruses could copy their RNA into DNA. Nobody had clear evidence that this was true and many people did not believe it, since the prevailing belief (called the “central dogma”) was that RNA was made from DNA (in a process called transcription), and not the other way around (Figure 2). Nonetheless, there was nothing impossible about copying RNA into DNA, because RNA and DNA are very similar molecules that are reproduced using similar mechanisms. In 1970, I decided to check the hypothesis that RNA might be copied “back” into DNA. To do that, I opened up RNA tumor viruses and added radioactive DNA precursors (Box 1). These are building blocks that allow us to detect the presence of any DNA made in a sample, because that DNA becomes radioactive. Within several days of experiments, I could show that the sample of RNA tumor viruses could make DNA [4].
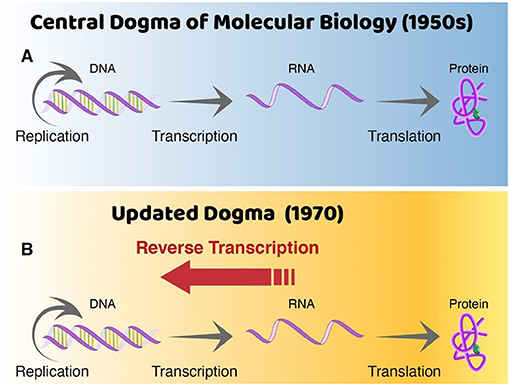
- Figure 2 - A change in dogma.
- (A) Prior to the discovery of reverse transcription, it was believed that RNA could be made from DNA through the process of transcription, but scientists generally did not believe that DNA could be made from RNA. (B) My discovery in 1970 showed that DNA can be made from RNA, by a process called reverse transcription. This finding changed one of the most basic ideas (dogma) in molecular biology. DNA replication, the process of making two identical DNA molecules from an original DNA molecule.
Box 1 - Finding DNA within RNA tumor viruses.
The process by which I used radioactive precursors to discover DNA went as follows: I bought ready-made nucleotides (the building blocks of DNA—A, T, C, and G), that were labeled with radioactive hydrogen. I put the virus and the labeled nucleotides in a test tube and used a detergent to break up the fatty membrane that covers the virus. The labeled nucleotides then contacted the RNA of the virus and the reverse transcriptase enzyme that copies RNA into DNA. I added some magnesium, which is necessary for the enzyme, and placed it in a 37°C bath. The enzyme built radioactive DNA from the labeled nucleotides. I used a strainer to separate the long DNA molecules from the remnants of the labeled nucleotides and, using a radiation detector, I found that the DNA molecules were radioactive! To prove that the molecule was indeed DNA, I used an enzyme called DNase that breaks DNA apart. When I added DNase to the reaction products and filtered it again using a strainer, I got no radioactive signal. That proved that the material I measured in the original reaction was definitely DNA.
My findings meant that RNA tumor viruses could make DNA out of their RNA genomes, because there was no other possible source of DNA in the experiment. I also showed that, if we eliminated the RNA from the samples, no DNA was found in them. This was a huge discovery of what is now called reverse transcription—a discovery for which I won, together with Renato Dulbecco and Howard Temin, a Nobel Prize in Physiology or Medicine in 1975—only 5 years after my experiments.
A New Enzyme That Converts RNA to DNA
When I discovered reverse transcription, it was known that RNA and DNA are copied by enzymes, so I knew that there was an enzyme in the virus particles. At that time, we did not know which enzyme converts RNA to DNA, nor did we understand the process used by that enzyme. My colleagues and I worked for 10 more years to discover the enzyme involved in reverse transcription and to decipher the complex mechanism by which it copied RNA into DNA. To do so, we developed a novel system, in which we could add known RNA molecules and examine what was made from them. This system allowed us to look at chemical reactions that no one had ever seen before. We found the enzyme that copies RNA to DNA and called it reverse transcriptase [5]. Several years after we found this enzyme, scientists from other research groups figured out its structure using an imaging method called X-ray crystallography (To learn more about the history of X-ray crystallography, see this Nobel Collection article.). Those scientists found that reverse transcriptase has a hand-like structure with “fingers,” a moveable “thumb” that can open and close, and a central area called the active site, where DNA is made from RNA (Figure 3).
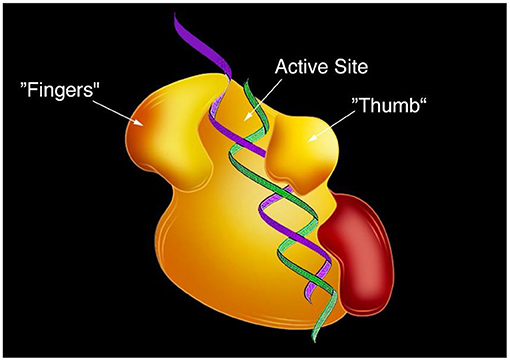
- Figure 3 - The structure of reverse transcriptase.
- Reverse transcriptase has a hand-like structure with “fingers” and a moveable “thumb” that can open and close. In its central area, it has an active site where RNA is made into DNA [Adapted from [6]].
New Treatments for Viral Infections and Cancer
Our discovery of reverse transcription and reverse transcriptase had major implications—both for our understanding of basic molecular processes in cells and for treatment of diseases. First, our discovery paved the way to understanding a type of viruses called retroviruses. Retroviruses are RNA viruses that use reverse transcription to create viral DNA from viral RNA. This viral DNA is then integrated (inserted) into the host cell’s DNA (using another viral enzyme called integrase), making it part of the host cell’s genetic material (Figure 4). After the viral DNA is integrated into the host’s DNA, the host produces viral proteins that assemble to form new virus particles.
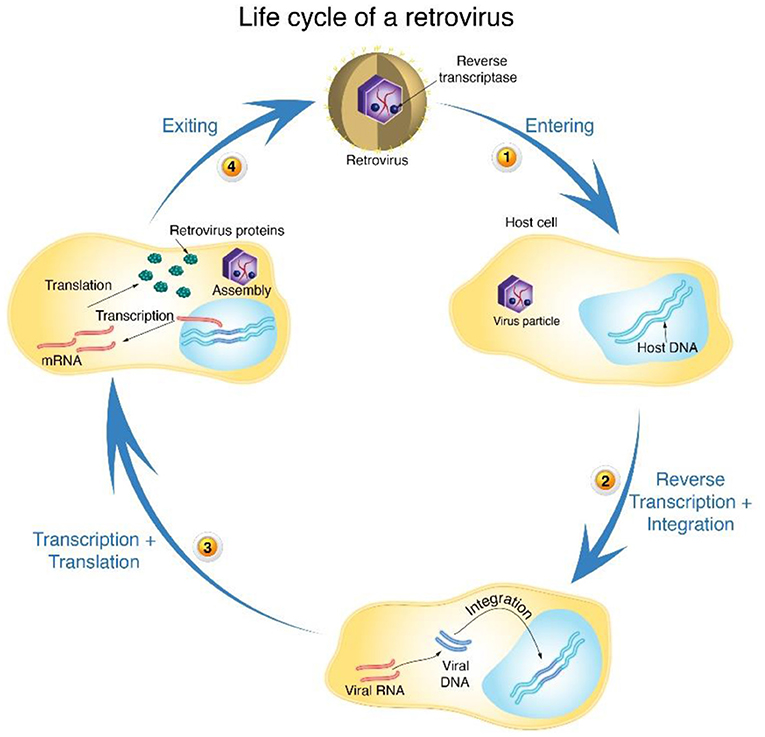
- Figure 4 - Life cycle of a retrovirus.
- (1) A retrovirus enters the host cell and sheds its outer envelope. (2) The retrovirus uses reverse transcription to create viral DNA from its RNA genome. The viral DNA is then integrated into the host cell’s genome, becoming part of the host’s DNA. (3) After integration, the host transcribes the viral DNA into mRNA and then translates the viral mRNA to produce viral proteins that are then assembled into new virus particles. (4) The new viruses go on to infect other cells (Image adapted from David Baltimore).
HIV, which causes AIDS, is one well-known and very problematic retrovirus (To learn more about HIV and AIDS, see this Nobel Collection article.). HIV was detected in 1983, about 2 years after AIDS became a global pandemic. HIV was detected based on its reverse transcriptase, and drugs were developed to treat AIDS. Had we not discovered reverse transcriptase in 1970, some 10 years before the emergence of AIDS, it would probably not have been possible to find the virus, and we would have spent a long time trying to understand why AIDS occurs. The relatively rapid discovery of HIV saved and improved the lives of many people who would have been less fortunate if AIDS had emerged before the discovery of reverse transcription.
Our discovery also played a key role in cancer treatment. Prior to our discovery, the way that RNA viruses cause cancer was a puzzle. In cancer, cells change from growing in a controlled way to growing out of control, sometimes into large masses called tumors in the body’s tissues or as rapidly growing cells in the blood, as in the case of leukemia. Since RNA is generally an unstable molecule, it did not make sense that RNA could cause a permanent change in a cell’s behavior. DNA, however, is a very stable molecule, and RNA tumor viruses were found to perform reverse transcription, copying their RNA into DNA, and basically turning themselves into genes of the cells they infect. The viruses then bring new genes into the cell and these genes make proteins. The viral proteins can override the host genes and force the cell to grow and divide continuously, making the infected cell a cancerous cell.
The connection between cancer and genes, which we discovered by studying RNA tumor viruses, turned out to be a very important process in the development of cancer. Our discovery provided one possibility of what goes wrong in cancerous cells, and it turned out to be true for multiple kinds of cancers, not only those caused by viruses. Our idea that cancer could be a genetic problem expanded the field of cancer research and triggered the discovery of life-saving drugs, including a “miracle drug” called Gleevec that blocks certain protein that signal cells to grow. The development of these drugs was based on the understanding that a specific gene caused a specific type of cancer. When we can inhibit the activity of a specific protein that is produced from that gene, we can inhibit the cancer.
Use in Biotechnology and Gene Therapy
Our discovery also helped to advance biotechnology, which often involves creating desired proteins for various applications, including drugs. One of the first things we asked after we discovered reverse transcriptase was: can it copy any RNA, or can it only copy viral RNA? It turned out that reverse transcriptase can copy any RNA into DNA, if we give it a little piece of DNA that is complementary to the RNA we want to copy, as a “starter.” So, this meant that we could turn any mRNA—the template for making a protein—into DNA, effectively making it into a gene. Once the mRNA was in the form of a gene, we could put that gene into cells (like bacteria) that could make lots of mRNA and then proteins from that gene. This ability to turn any mRNA into DNA and then make many copies of a desired protein was revolutionary for the biotechnology industry and led to the development of many new drugs.
Retroviruses are commonly used in gene therapy, as a tool for curing genetic diseases [7]. One very successful example of retroviral gene therapy is for the treatment of bubble baby disease, officially called severe combined immunodeficiency (SCID). These babies have no functioning immune system, so any infection is lethal for them. In the past, these babies had to live in plastic bubbles that separated them from the outside environment so they would not get sick. Nowadays, novel retroviral gene therapies that replace the babies’ damaged genes with functioning genes can restore their immune systems and allow them to live normal lives.
I was not originally focused on studying diseases and trying to cure people. I was initially interested only in the basic science. I was curious about viruses and wanted to understand them and how they affected their hosts. But the way it turned out, the new knowledge I helped uncover had, and still has, great implications for biotechnology and certain areas of medicine. This is one example of a recurring theme in science: life-improving and even lifesaving applications often spring from new scientific knowledge.
Recommendations for Young Minds
When I work with students, I encourage them to be as independent as possible—to think independently about problems and solutions and to work on those problems in labs that support them in their quest to be independent. It is often difficult for young students to work independently, but they must do this because science is based on the work and knowledge of individual scientists. Nowadays, we are very focused on cooperation and collaborative work, and that is very important. But ultimately, it is the imagination, knowledge, and hard work of individual scientists that lead to discoveries like reverse transcriptase and so many others that are made every day. So, I encourage my students and I encourage you to choose a career path that gives you independence as early as possible, and that allows you to find your own way in science (Figure 5). That means you do not just mimic the ways that your supervisors and advisors do science, but instead you find your unique way of doing things. To conduct a successful study, you need an in-depth understanding of the topic you are focusing on, such as a particular organism or disease. When you first start out in biology, where things are complicated and require many different skills—you need to have in-depth understanding and focus only on one or two defined problems, because that will allow you to deepen your knowledge. Later in your career, you might choose to take a broader look at what is going in other fields of biology and other areas in science but, in the earliest stages of your career, focus and depth are very important.

- Figure 5 - Recommendations for young minds.
- I encourage all of you who are interested in a career in science to be independent and find your own way.
My greatest enjoyment as a scientist comes from discovering new things through trying to solve biological problems. I think that is the most rewarding life a person can possibly live, and I encourage people to move in that direction, if their minds work that way. Many people do not want to spend their time solving problems and that is fine. The most important thing is to figure out what gives you pleasure and then go into it full-time. It is not easy being a scientist, nor is it easy doing anything else with full commitment and enormous depth. But, once you get into that depth, it gives you rewards that you may never achieve any other way.
Glossary
Virus: ↑ A tiny particle that infects living cells and uses the cells’ machinery to make more viruses. Viruses can infect all life forms.
Messenger RNA (mRNA): ↑ A type of RNA that carries the instructions for making proteins and moves from the nucleus to the protein factory of the cell (ribosome).
Enzymes: ↑ Proteins that control the speed of chemical reactions in living cells.
RNA Tumor Viruses: ↑ RNA viruses that cause tumor by integrating their genome into the host genome.
Reverse Transcriptase: ↑ An enzyme that performs reverse transcription.
Reverse Transcription: ↑ The process by which DNA is copied from RNA—the “reverse” of normal transcription, in which RNA is copied from DNA.
Retroviruses: ↑ Viruses that make DNA by copying their RNA genome using reverse transcription.
Biotechnology: ↑ An industrial field that uses biological processes to develop products.
Acknowledgments
I wish to thank Noa Segev for conducting the interview which served as the basis for this paper and for co-authoring the paper, and Zehava Cohen for providing the figures.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
[1] ↑ Baltimore, D. 1971. Expression of animal virus genomes. Bacteriol. Rev. 35:235–41.
[2] ↑ Koonin, E. V., Krupovic, M., and Agol, V. I. 2021. The Baltimore classification of viruses 50 years later: how does it stand in the light of virus evolution? Microbiol. Mol. Biol. Rev. 85:e00053-21. doi: 10.1128/MMBR.00053-21
[3] ↑ Baltimore, D., and Franklin, R. M. 1962. The effect of Mengovirus infection on the activity of the DNA-dependent RNA polymerase of L-cells. Proc. Natl. Acad. Sci. 48:1383–90.
[4] ↑ Baltimore, D. 1970. Viral RNA-dependent DNA polymerase: RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 226:1209–11.
[5] ↑ Coffin, J. M., and Fan, H. 2016. The discovery of reverse transcriptase. Annu. Rev. Virol. 3:29–51. doi: 10.1146/annurev-virology-110615-035556
[6] ↑ Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. 2002. Site-Specific Recombination. In Molecular Biology of the Cell. 4th ed. New York, NY: Garland Science.
[7] ↑ Anson, D. S. 2004. The use of retroviral vectors for gene therapy-what are the risks? A review of retroviral pathogenesis and its relevance to retroviral vector-mediated gene delivery. Genet. Vacc. Ther. 2:1–13. doi: 10.1186/1479-0556-2-9
