Abstract
This article is based on an interview between the two authors.
Structural biology is a field that seeks to find the structures of all the components that make up living things—from molecules that exist in humans and other animals, through molecules present in tiny microorganisms, to the molecules that make up plants. To determine these structures, structural biologists use sophisticated imaging techniques that are becoming more and more accurate at “seeing”, or determining the structure of smaller and more diverse molecules. Electron cryomicroscopy is one very advanced and powerful imaging technique. In this technique, electrons are sent through frozen specimens to determine the structures of single molecules, at magnifications that are enough to see atoms. These images are taking us one step further toward understanding the structure and function of the basic building blocks of life. In this article, we will tell you about the developments that led to what is called “the resolution revolution” in electron cryomicroscopy, which Dr. Henderson was part of and that eventually allowed him to share the Nobel Prize in Chemistry in 2017.
Dr. Richard Henderson won the Nobel Prize in Chemistry in 2017, jointly with Prof. Jacques Dubochet and Prof. Joachim Frank, for developing cryo-electron microscopy for the high-resolution structure determination of biomolecules in solution.
When Looks Matter: Discovering the Structures of Biological Molecules
Living things contain many important structures and processes. In the human body, for example, we know there are organs, which are made from cells, and within these cells are many organelles and molecules that perform all the functions necessary to maintain life—such as producing energy, getting rid of waste, moving, and defending against harmful factors (Figure 1). To understand how living things work and to possibly improve their lives, we need to know which structures are present in a biological system and which activities those structures perform. Structural biology is the field that seeks to observe the structures of biological components. In the past, scientists would first look for specific activities that we knew were happening in a biological system, such as the conversion of one source of energy to another, storable type of energy. After we found the activity, we would identify the molecules that took part in it—usually proteins and enzymes, and only then determine the structures of those molecules.
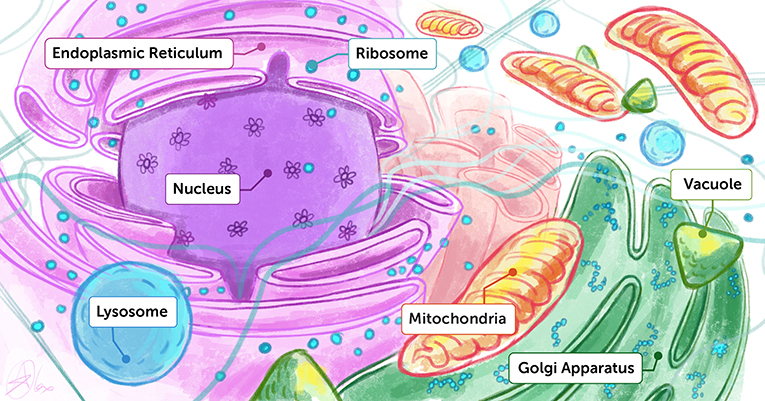
- Figure 1 - Artist’s interpretation of the inside of a cell.
- You can think of the inside of a cell like a dense playground containing many different molecules and organelles, each performing their unique functions. To understand how life works, we want to know both the structures and the functions of each of these biological building blocks.
In 2000, the world experienced a revolution in the understanding of genetic information (DNA) the information stored in our cells, which we inherit from our parents. Scientists put together the first complete “set of instructions" (called the sequence of DNA bases) of the whole package of human genetic information. Since then, rather than looking for an activity, then finding a molecule, and then determining the structure of that molecule, structural biologists can use the genetic information, which tells them about all the enzymes and proteins in the body. In 2000, when the human genome was determined, about 80% of the molecules coded for in human DNA had not even been discovered! This opened up a whole new pathway in structural biology—we could now find the structures of molecules without having to know their functions first. How do we find the structures of these molecules? Using tiny particles called electrons!
Electrons and Electron Microscopy
Electrons are tiny electrically charged particles present in every atom. The movement of electrons is what creates electricity, and electrons are also the source of light and other forms of electromagnetic radiation such as X-rays. Can you believe that, until 1895, no one even knew electrons existed? In that year, electrons were first identified and named by JJ Thompson, a scientist in the physics department at the University of Cambridge in the United Kingdom. Thirty years later, in 1935, GP Thompson (the son of JJ Thompson), showed that electrons behave as both particles and waves—they have a frequency and a wavelength, just like other waves do. JJ and GP Thompson were both awarded Nobel Prizes: for the discovery of the electron as a particle and for the discovery of the electron as a wave.
Shortly after that, scientists realized that if electrons behave like waves, in some sense they must behave like light—because light can also behave like a wave. So, scientists thought that maybe they could use electrons to illuminate tiny specimens they wanted to look at, the same way we can look at an object with our eyes, with a camera, or with a regular microscope—but using electrons instead of visible light. This was the start of electron microscopy. Electron microscopy is a very powerful imaging technique. Electrons have a very short wavelength, about 100,000 times shorter than the wavelength of light. You can think of the wavelength as a “zooming” parameter—the smaller the wavelength, the more we can “zoom in” on our specimen. This means that pictures taken with electrons have a very high level of detail, which is called a high resolution. Due to its high resolution, electron microscopy can be used to discover the structures of tiny molecules in a way that was not possible previously.
How Does Electron Microscopy Work?
In electron microscopy, an energetic beam of electrons emerges from an electron source and passes through the specimen being studied (Figure 2A). When electrons pass through the specimen, they interact with its atoms and get diffracted, or thrown off course, in a way that is specific to the arrangement of atoms they encounter. In this way, the electrons “pick up” the structure of the material as they pass through it. After the electrons are diffracted, they are collected and focused using lenses (much like the lenses inside a camera), and then they are recorded by an electron detector. At this stage, scientists have an image of the electrons that were diffracted from the specimen, and they must convert it to an image of the specimen itself. This conversion is based on simple physics that describes the relationship between a measured object and the resulting image. The conversion depends on many factors, including the wavelength of the electrons and the lenses that are used; but that is all taken care of by microscopy experts.
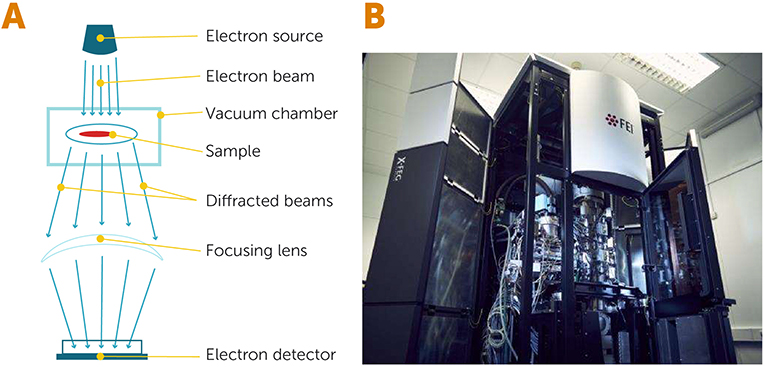
- Figure 2 - Electron microscopy.
- (A) In an electron microscope, a source of electrons releases a beam of hot, energetic electrons that passes through a specimen, which is located inside a vacuum chamber. When the electrons interact with the specimen, they get diffracted (scattered) and are then collected and focused by special lenses before being detected by an electron detector. (B) An electron microscope from the University of Cambridge (United Kingdom), which allows scientists to image frozen biological specimens (Image credit: University of Cambridge).
Challenges in Electron Microscopy
Even though electrons can help us get remarkable images of molecules, there are major challenges to be overcome when using them to image biological molecules. First, as quantum physics tells us, individual electrons are not “logical”—when you ask them a question (for example, what happens when they meet a specific molecule), they do not return a definite answer like a human would. Instead, they have a certain probability (likelihood) of participating in each possible outcome. In the electron world, everything that could happen did happen, with a certain probability for each option. This means that scientists must collect many answers from many electrons, and combine the information intelligently to get the overall answer. To accomplish this, we illuminate the specimen with millions of electrons and use the overall average of their properties to obtain a sensible answer.
Second, the electrons can damage the specimen, because they are very energetic and must pass all the way through the specimen to arrive at the detector. Their temperatures can get to about two thousand million°C—for comparison, water boils at 100°C! These super-energetic electrons, and any other type of energetic radiation, can pull electrons out of the specimen’s molecules. This changes the shapes and properties of the specimen’s molecules, because biological molecules are relatively delicate. Therefore, it is difficult for scientists get enough information about the structure of a single biological molecule before it is destroyed. One way to address this challenge is to take images of many separate, identical molecules—at least 500, in the case of electron microscopy of biological molecules—and average the images to get the structure of a typical molecule. Another way we can deal with this challenge is to cool the specimen in a special way that makes it more resilient to electron damage—this will be described in the next section.
Yet another challenge comes from the fact that electrons are diffracted as soon as they are near any atoms. This means that there must be a completely clear pathway between the electron source and the specimen, so that the electrons will arrive at the desired molecules and not be scattered due to other molecules, even oxygen and nitrogen in the air, that are in the way. In other words, scientists must create a vacuum around the specimen in the electron microscope. Since biological molecules are always in a water-containing solutions (think, for example, about the molecules in your blood), the problem is that water evaporates in a vacuum, and the specimen dries up. This drying often damages biological molecules in the specimen. Overcoming this challenge required structural biologists to use their creativity to take advantage of the unique properties of water.
Can Water Stay Liquid When It Is Very Cold?
Here is a really nice experiment you can try at home to understand one of the unique properties of water (Figure 3). Take an empty jar with a lid and fill it full of water. Try to make sure there are no air bubbles in the jar by screwing the lid on tightly under water and put it in the freezer. Leave the jar in the freezer for a day—by then the temperature of the water will go down to -10°C or -20°C (Water normally becomes ice at 0°C). The next day, take the jar out of the freezer and take a look—has the water turned into solid ice, or did it stay liquid?
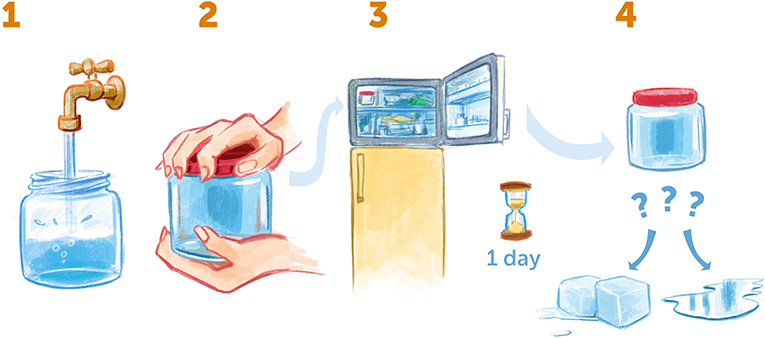
- Figure 3 - Supercooling water at home.
- (1) Take an empty jar, fill it up completely with water, and make sure there are no air bubbles inside. (2) Seal the jar tightly and (3) put it in the freezer for a day. (4) Then, take the jar out. Is the water frozen or still liquid? If it is still liquid, you have created supercooled water!
Most often, you will find that the water is still liquid—it has been supercooled, meaning that it cooled down to a temperature lower than its freezing temperature (0°C), but without turning into ice. In our experiments, we want to cool water even further, to below -170°C, because at this temperature it becomes very still and stable. We also want to avoid creating ice crystals, because they interfere with our measurements. To accomplish this, we must use a special cooling method developed in the lab of Jacques Dubochet, who shared the Nobel Prize in Chemistry with me, Richard Henderson, and Joachim Frank in 2017. In this method, we use very cold liquid ethane or propane (substances found in natural gas, made from carbon and hydrogen), cooled to -185°C. We then plunge a very thin film of water into the cooled ethane/propane liquid, and it cools down so quickly—in about 1/1,000 of a second—that it has no time to form organized ice crystals and stays in the unorganized, liquid form [1] that we call amorphous ice. In this way, we get supercooled water.
The Magic Formula of Hot Electrons and Cold Specimens
It turns out that supercooled thin films of water are great for suspending the biological molecules we want to image with electron microscopes. When we add this cooling step to the imaging process, this is called electron cryomicroscopy (“cryo” is short for “cryogenic,” which refers to cooling). Electron cryomicroscopy allows us to deal with two of the challenges mentioned above: it stabilizes the specimen, making it more resilient to damage by high-energy electrons, and it allows biological molecules to be in their natural watery environment, without evaporation of water due to vacuum. There is one more important advantage: unlike most other liquids, water expands when it is cooled below 4°C. This feature of water helps biological molecules to remain undamaged when they are in supercooled water. If water contracted when cooled, it would squeeze the molecules and possibly break them.
This rather simple yet highly effective electron cryomicroscopy imaging method allows us to image biological molecules at a resolution that was previously unattainable. This is why it is sometimes referred to as the “resolution revolution”.
Figure 4 shows examples of beautiful images obtained using electron cryomicroscopy. We are dealing with extremely tiny scales-10’s of nanometers, which is less than 1/1,000 the width of a human hair! Hopefully you are beginning to appreciate the wonder of electron cryomicroscopy.
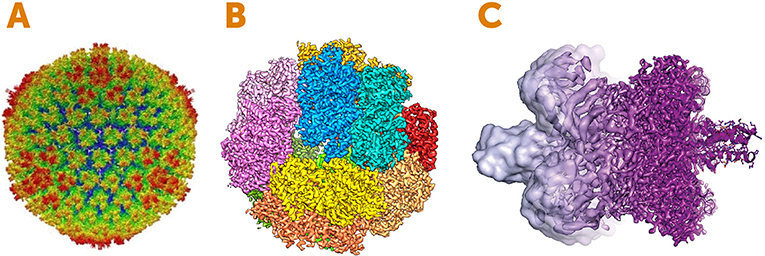
- Figure 4 - Images created using electron cryomicroscopy.
- (A) Structure of a disease-causing virus called Adenovirus. The image shows the outer surface called the capsid, which is the protein shell that wraps the virus’s genetic material. The colors represent the distance from the center of the sphere: red is the furthest from the center and blue is the nearest. (B) An enzyme involved in energy generation in microbes. The colors represent the individual subunits (pieces) of the enzyme. (C) An example of how the resolution of electron cryomicroscopy has improved between 2013 (left, light purple) and 2017 (right, dark purple). Image Credits: (A) adapted from [2]; (B) adapted from [3]; (C) Martin Högbom, Stockholm University, based on an image by V. Falconieri.
What Does the Future Hold for Electron Cryomicroscopy?
Electrons are the best particles to use for imaging tiny biological molecules. To give you a sense of just how good they are, let us compare them to two other commonly used particles: X-ray photons (similar to light particles but with short wavelengths), and neutrons (particles from the nuclei of atoms). When we calculate the amount of information we receive from using a certain particle compared with the damage that particle creates in the specimen, we get a measure of how good the particle is for imaging. Using that calculation, electrons are 1,000 times better than X-rays, and three times better than neutrons! That is why my colleagues and I started using electrons instead of other particles many years ago. Nowadays, electron cryomicroscopy has become so successful that the number of structural biologists using it is high and increasing rapidly.
There are still significant improvements to be made in electron cryomicroscopy. One is improving the electron detectors, which are still not large or efficient enough and require us to use many more electrons than theoretically required. It would also be helpful to minimize the motion of the specimen (both the water molecules and the biological molecules) when it contacts the electron beam [4, 5]. We believe that, in about 5 years’ time, there will be significant advances to address these challenges. We will then have an even more powerful tool that will allow us better to understand many biological questions, such as how life works and how it reproduces. The information we acquire might help us to maintain the health of people, animals, and plants. We can expect a very bright future for electron cryomicroscopy!
Recommendations for Young Minds
I, Richard, want to share some practical advice that I have followed throughout my career. It comes from the writing of Peter Medawar, who won a Nobel Prize in Physiology or Medicine in 1960. After winning the Nobel Prize, Peter Medawar published two books called “The Art of the Soluble,” and “Advice to a Young Scientist.” In his books, he said that there are many interesting things in science and life, and that we should be interested in everything. But we should also pick something that we are particularly interested in to work on. Moreover, he said that scientists should work on something that will successfully produce new knowledge soon—not in 100 years, because that is beyond a scientist’s lifetime. His idea was that science is the art of the soluble, meaning problems that can be solved. Scientists should do experiments that work now, with current techniques.
When I was a young student of physics, I wondered where physics was going, and I remember making a list of all the interesting topics of the future. There was fusion research, which involves producing unlimited power from hydrogen fusion. Then there was high-energy particle physics, which led to the discovery of new particles, including the Higgs boson and more. There was solid-state physics, which advanced the computer industry and the development of the microchips that fuel computers. Biophysics, astrophysics, cosmology, black holes, and neutron stars, to name just a few, were other interesting topics (Figure 5). If I had picked any of those topics to study, they would have been equally interesting and exciting. So, if you decide to go into science, you must pick something that you are interested in, so that you will keep studying and working on it without anybody forcing you to do so. When you are interested and self-motivated, if you run into difficulties. it will not bother you too much—you will just take it as a challenge and move on. Once you have chosen an interesting topic, before you actually head off in that direction it is best to find out as much as you can about the various activities that you could pursue to study the topic. If, after 6 months or a year of your best effort, it turns out that your idea was not so good, do not hesitate to think again and find a new direction.

- Figure 5 - Choosing a scientific field.
- When I was a physics student, there were many interesting topics I could choose from. In hindsight, I realize that all of them would have been equally interesting for me. When you chose a field, make sure to choose something that you find interesting and that will keep you motivated over a long period of time.
Today, science is moving very rapidly compared to the past. Only a 100 years ago, we did not even know about the existence of X-rays and electrons, and now we have information about the whole human genome, we have sophisticated methods to work with DNA, and we can figure out the structure of virtually anything we want. The next 100 years will be a very good time to be alive—and to be a scientist. Enjoy your life and invest yourself in whatever is most interesting to you!
Additional Materials
Richard Henderson Nobel Lecture 2017.
Electron Cryomicroscopy—Richard Henderson at Serious Science.
Explainer: What is cryo-electron microscopy (Chemistry World).
Glossary
Structural Biology: ↑ A field of biology characterizes the structures of the components that make up living things.
Protein: ↑ A tiny biological machine that performs many essential roles in the body.
Enzyme: ↑ A biological molecule that speeds up chemical reactions in the body.
Genetic Information (DNA): ↑ Information passed on from parents to their offspring, that determines the characteristics and behavior of an organism.
Frequency: ↑ The number of times a wave repeats itself in one second.
Wavelength: ↑ The distance over which the shape of a wave repeats itself.
Electron Microscopy: ↑ A technique that uses electrons to image small structures, including biological molecules.
Diffraction: ↑ The scattering of a wave by an object; for example the scattering of electrons by a specimen.
Supercooled: ↑ Cooled below the freezing temperature while staying liquid (i.e., without crystalizing).
Electron Cryomicroscopy: ↑ A technique that uses electrons to image biological molecules that are in water and are cooled to low temperatures very rapidly.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Alex Bernstein for providing the figures and Susan Debad for copyediting the manuscript.
References
[1] ↑ Dubochet, J., Lepault, J., Freeman, R., Berriman, J. A., and Homo, J.-C. 1982. Electron microscopy of frozen water and aqueous solutions. J. Microscopy 128:219–37. doi: 10.1111/j.1365-2818.1982.tb04625.x
[2] ↑ Liu, H., Jin, L., Koh, S. B. S., Atanasov, I., Schein, S., Wu, L., et al. 2010. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329:1038–43. doi: 10.1126/science.1187433
[3] ↑ Allegretti, M., Mills, D. J., McMullan, G., Kühlbrandt, W., and Vonck, J. 2014. Atomic model of the F420-reducing [NiFe] hydrogenase by electron cryo-microscopy using a direct electron detector. Elife 3:e01963. doi: 10.7554/eLife.01963
[4] ↑ Vinothkumar, K. R., and Henderson, R. 2016. Single particle electron cryomicroscopy: Trends, issues and future perspective. Q. Rev. Biophys. 49:e13. doi: 10.1017/S0033583516000068
[5] ↑ Henderson, R. 2015. Overview and future of single particle electron cryomicroscopy. Archiv. Biochem. Biophys. 581:19–24. doi: 10.1016/j.abb.2015.02.036
