Abstract
Proteins are small biological machines that work in our bodies as well as in the bodies of all animals, plants, viruses, and bacteria. They are responsible for a wide range of vital activities. Proteins are synthesized based on instructions contained in the genetic code, within their DNA, by a cellular organelle called ribosomes. The ribosome assembles proteins very quickly and accurately. Most of my scientific research is devoted to understanding the function of the ribosome, based on its structure, as we determined it. In this article, I describe the major findings of my research, some important applications of these findings, and the challenges I have faced along the way.
Professor Ada Yonath was awarded the Nobel Prize in Chemistry in 2009, jointly with Prof. Venkatraman Ramakrishnan and Prof. Thomas A. Steitz, for studies of the structure and function of the ribosome.
From DNA to Proteins
Each living cell contains a genetic code, which is the instructions for the creation of the cell components and the entire living organism. This substance is inherited from parents to their offspring. The genetic material is called DNA and can be considered a cookbook that contains all the information necessary for the creation and functioning of the living organism. One of the most important components of DNA are genes, which contain instructions for the synthesis of the proteins—the “machinery” of the cells and of the entire living body. Proteins perform many actions that are important for the functioning of the body.
All natural proteins consist of a combination of about 20 building blocks called amino acids. To form a protein, the amino acids are joined together like beads on a chain. This connection is called a peptide bond, and a medium-sized protein contains between 150 and 500 amino acids linked together. Each such long chain of amino acids folds into a unique, functional three-dimensional structure determined by the sequence of the amino acids that make it up. The protein structure is precisely tailored to execute the function that the protein is intended to perform (Figure 1).
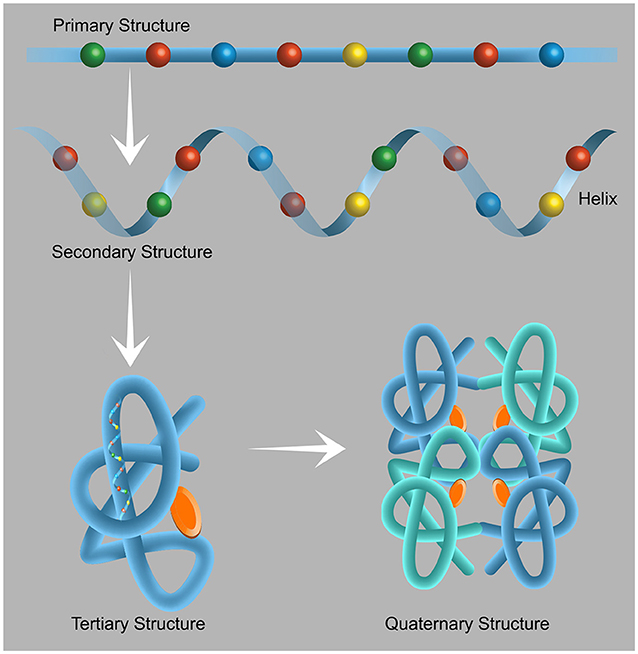
- Figure 1 - The structure of proteins.
- Proteins are made up of 20 building blocks called amino acids. When the amino acids are joined together, they form a chain called the primary structure of the protein. Each amino acid has its unique chemical properties that are necessary for the protein to function. Therefore, the sequence in which the amino acids are ordered is extremely important. Each of the protein chains folds according to the properties of its components to form the secondary structure, which can take a variety of forms, including that of a helix. The tertiary structure is the three-dimensional structure of the protein, i.e., the arrangement of the helices and the chains connecting the helices that enables the specific execution of the specific functions of that protein. Some proteins are active in the form of clusters—the arrangement of proteins in a cluster is called a quaternary structure. These clusters consist of at least two protein components bound together in a way that enables their activity. Figure adapted from here.
Protein production in cells occurs in several stages (Figure 2). In the first stage, a part of the DNA which contains the genetic material, is copied into messenger RNA (mRNA) in a process called transcription. An mRNA molecule is a “copy” of the genetic information that allows the genetic material to be transferred inside the cell without “endangering” the original code. In the next stage, the mRNA reaches the ribosome, the cell’s “factory” for protein production. The ribosome “reads” the instructions from the mRNA and prepares the desired protein by forming a chain of amino acids joined to each other by peptide bonds. After exiting from the ribosome, the protein chains are folded into the proper three-dimensional structure required for the protein to function. This process of protein production in the ribosome is called translation. The rest of this article will focus on how the translation process occurs in the ribosome.
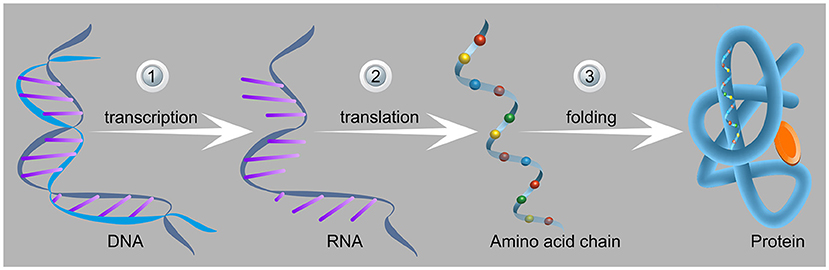
- Figure 2 - Stages of protein production in cells.
- (1) In the first stage, the part of the DNA containing nucleotides with the sequence of the protein (left) is transcribed to generate the messenger RNA. (2) In the second stage, the mRNA and the amino acids reach the ribosome and create the amino acid chain (namely the protein) that leaves the ribosome through a protected tunnel. (3) After exiting from the ribosome, the chain folds into the three-dimensional structure of the protein (right). This unique structure is appropriate for the function of the protein produced (one of the many different functions of the various proteins in the body). Image adapted from here.
The Ribosome
The translation of a new protein is a complicated process. The “recipe” for proteins is encoded in the genetic code (DNA and mRNA), in a genetic “language” of four letters that is translated into a “language” of amino acids that contains about 20 different building blocks—the amino acids. You are probably wondering, “How is this possible?” The process is called translation, and the name suggests the process. The solution to this problem is that the ribosome reads the genetic code as a series of three-letter combinations of nucleotides called codons. Each codon corresponds to a particular amino acid (some amino acids have more than one codon. Sometimes there are up to four different codons that are translated into the same amino acid) or a stop signal. The ribosome is responsible for the translation, namely creates the connections between the amino acids into the nascent protein.
The ribosome is a cellular organelle composed of two subunits: the small ribosomal subunit and the large ribosomal subunit (Figure 3), each of which has a specific function. The small ribosomal subunit is responsible for reading the genetic code from the mRNA and matches between the bound mRNA codon and the corresponding anticodon on the transfer RNA (tRNA). The site where the genetic code is read is called the decoding center. The large ribosomal subunit is responsible for catalyzing the new peptide bond between the amino acids and for passing the synthesized protein through the ribosome into the Nascent Peptide Exit Tunnel (NPET). In the catalytic center of the ribosome, the peptidyl transferase center (PTC), the amino acids bind to each other, and a new peptide bond is formed. The PTC allows the bonding of the two amino acids as within it, they are situated in close proximity and in the proper orientation to create a new peptide bond. After the new bond is formed, the tRNA holding the nascent protein moves together with the mRNA to the “next station” across the ribosome, ready to form a new peptide bond. The nascent protein chain passes through the large ribosomal subunit within the NPET and emerges at the other end of the ribosome.
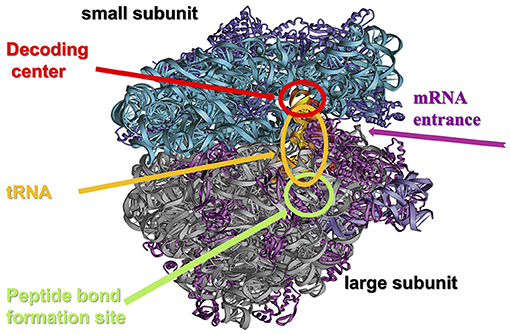
- Figure 3 - The structure of the ribosome.
- The ribosome consists of a few dozen proteins (purple and magenta) and chains of ribosomal RNA (turquoise and gray), organized into two subunits: the small ribosomal subunit (turquoise and purple) and the large ribosomal subunit (magenta and gray). The small subunit contains the site where the genetic material is read, called the decoding center, into which the mRNA molecule enters and in which it binds to the transfer RNA (tRNA) according to the genetic code, represented by three letters (see Figure 4). The large ribosomal subunit contains a site called the peptidyl transferase center (PTC), where the amino acids, carried by tRNA, bind to each other. The two subunits of the ribosome come together during the assembly of a new protein and are separated at the end of protein assembly (Figure credit: Ada Yonath).
The small and large ribosomal subunits are bound to each other during protein assembly and detach from each other when protein translation is complete. Each ribosomal subunit consists of proteins and very long RNA chains called ribosomal RNA (rRNA) (Figure 3) [1]. There are many ribosomes in each living cell—from tens of thousands in bacteria, for example, to millions in human cells. The work of ribosomes requires a lot of energy—more than 60% of the total energy used by the cell [2]! This highlights the importance of the ribosome within the cell, as it is the factory that produces the proteins required for all cellular activities. The ribosome works at an amazing speed: a bacterial ribosome can produce a protein of average size within a few seconds to a minute.
Ribosome translation of an mRNA molecule occurs in three stages: Initiation, Elongation, and Termination: during initiation, an mRNA binds to the small ribosomal subunit of the ribosome. Then a tRNA carrying the amino acid methionine binds to the start codon of the mRNA sequence. The start codon in all mRNA molecules has the sequence AUG, which codes for methionine. Initiation factors join the small ribosomal subunit, mRNA, and tRNA complex, and the initiation complex is formed. Next, the large ribosomal subunit binds and forms the translation complex, i.e., the active ribosome. During the elongation stage, the ribosome translates every codon in turn. For each amino acid, there is a separate and unique tRNA that binds to the ribosome and carries the amino acid with it. As the ribosome moves across the mRNA, new tRNA molecules loaded with the corresponding amino acid keep binding and moving between the three active sites of the ribosome: the Aminoacyl site (A site), Peptidyl site (P site), and the Exit site (E site) (Figure 4). Then the next tRNA carries an amino acid, enters the ribosome, binds to the A site, a peptide bond is being formed between the amino acids bound to the A site and the P site, and the nascent protein chain moves on within the NPET. Later, the tRNA from the P site moves to the E site and is released from the ribosome. The tRNA from the A site moves to the P site, and a new tRNA binds to the A site. Each time a new amino acid arrives, it binds to the previous amino acid by a peptide bond. In this way, the nascent protein chain is formed [3]. The growing peptide chain passes through the ribosome through a Nascent Peptide Exit Tunnel (NPET) and emerges from the ribosome. Elongation continues as long as the mRNA advances in the ribosome and presents new codons. Termination occurs when the ribosome reaches a stop codon. A stop codon is a codon on the mRNA for which there are no tRNA molecules that can recognize it. When the translation is complete, the two subunits of the ribosome are separated and the nascent protein exits and folds into its three-dimensional structure, alone or with helpers (chaperones). In this way, the new protein is released from the ribosome and can perform its function in the cell (a video describing this process, can be found here).
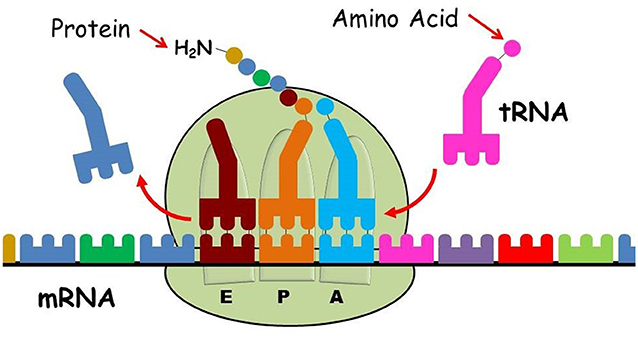
- Figure 4 - Protein production in the ribosome.
- The ribosome is composed of two distinct and separate subunits: the small ribosomal subunit (bottom) and the large ribosomal subunit (top). The small ribosomal subunit decodes the mRNA (multicolored chain) that contains the instructions for assembling the protein. The large ribosomal subunit catalyzes the formation of peptide bonds and synthesizes the nascent protein chain according to the instructions of the mRNA as follows: the first transfer RNA (tRNA) loaded with the corresponding amino acid (colored circle) binds to the peptidyl site (P site), then the next loaded tRNA binds to the aminoacyl site (A site) of the ribosome. A new peptide bond is formed between the amino acids bound to the tRNA at the peptidyl site (P site) and the A site (pale blue and orange circles in the A and P sites). The tRNA from the P site is transferred to the exit site (E site) (dark red), the tRNA from the A site is transferred to the P site with the nascent protein chain bound to it, and a new tRNA loaded with the next amino acid can bind the A site. At the end of the process, the amino acid chain is released from the ribosome and folded into the functional protein (Image credit: Ada Yonath’s modified Google figure).
How did we discover the structure of the ribosome? We developed a unique method to do this, which we will discuss in the next section.
Crystallography Inspired by Polar Bears
We have deciphered the structure and function of the ribosome using a method called Crystallography. This method enables us to study crystallized materials. A crystal is a structure consisting of units with internal symmetry that repeat in space in an orderly fashion. The simplest and smallest repeating unit in a crystal is called a “unit cell”. A unit cell of table salt is shown in Figure 5A. Each unit cell in a crystal has a fixed measure of length, height, width, and fixed angles between its feces. In a crystal, the unit cells are arranged periodically in three dimensions, side by side, like floor tiles. In crystallography, the crystal is irradiated with a uniform and very concentrated X-ray beam (Figure 5B). The atoms presented in each unit cell of the crystal diffract the X-ray beam to a few specific directions, determined by the physical law, called Bragg’s law, and their properties and positions, once the X-ray beam hits them. During irradiation, images of the scattering of the beam are captured and collected on a screen (Figure 5B, screen at right) or detector. Using these diffraction patterns by mathematical procedures called Fourier equations, we can decipher the three-dimensional structure of the unit cell.
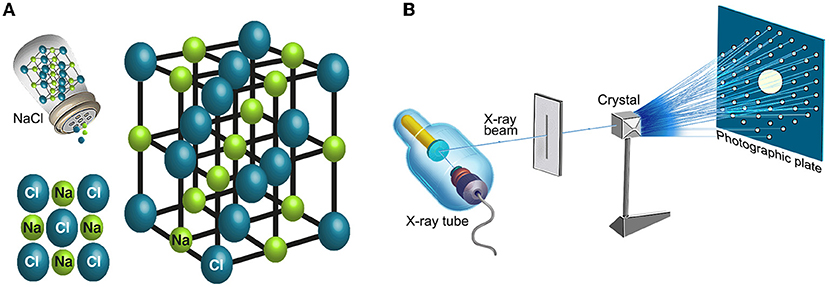
- Figure 5 - Deciphering the structure of a crystal using crystallography.
- (A) An example of the atomic arrangement in a crystal known from our daily lives—table salt. Table salt consists of two ions: sodium cation (a positive ion—in green) and chloride anion (a negative ion—in blue), arranged periodically in unit cells that repeat in all directions of space. (B) Determining the structure of a crystal using crystallography: the crystal is irradiated with a uniform and concentrated X-ray beam. Due to the arrangement of the atoms that make up the crystal, the X-ray beam is diffracted after hitting the crystal. During the experiment, the crystal is irradiated from several angles, and the strength and position of the dots of the image appearing on the screen are measured (right). Using the image of the diffracted beam, we can decipher the three-dimensional structure of the crystal.
The first requirement for deciphering structure using X-rays is the preparation of crystals. Some materials, such as sugar, table salt and quartz stone, are natural crystals—these crystals are stable and solid. But most materials and structures in nature and in the body, such as proteins (e.g., hemoglobin) or their complexes, like ribosomes, are not naturally crystallized. Therefore, the first challenge in our research was to find conditions to crystallize the multi-components complicated and flexible ribosomes—a task previously considered impossible, since many famous scientists had already tried and failed. For a long time, we could not crystallize ribosomes because of their extremely complicated structure. Due to their functional requirements in the body, they have motor abilities—and tend to disassemble outside the cell, making their crystallization even more difficult.
One of the many important breakthroughs we made while trying to crystallize the ribosome was inspired by polar bears. It turned out that during their winter hibernation, a large portion of the bears’ ribosomes are maintained in a dense, almost periodic structure throughout the winter, until the bears awaken from their sleep in the spring. We assumed that this happens so that the bears’ ribosomes do not fall apart when they are inactive over the winter—so that they can still function in the spring. This told us that, under stress or extreme conditions, the ribosomes tend to arrange in an almost periodic density, and therefore we chose to produce crystals from ribosomes of bacteria living under extreme stress conditions in the dead sea. Indeed, through intense work and after trying countless different conditions, we managed to get stable crystals of active ribosomes.
After producing the crystals, we knew we would face another difficulty. The X-ray beam that we had to use to decipher the structure of the ribosome was of very high energy (produced in giant accelerators, e.g., un Grenoble, Stanford or APS) that affects the chemical bonds between the atoms of the ribosome crystals and causes migrating atoms. This means that as we irradiate the crystal to collect data about its structure, we are destroying it. In practice, therefore, we would not be able to measure more than a few percent of the information required to determine the ribosome’s structure. We knew we had to find a way to collect enough data from the crystallized ribosome before it was destroyed. Hence, we tried to make crystals that would last longer so that we could make more measurements. We developed a method to reduce the damage from the X-ray radiation by rapidly freezing the crystals at very low temperatures (around -200 degrees Celsius, by using liquid nitrogen), under conditions that would prevent the formation of ice crystals in the liquid surrounding the crystallized ribosome, aiming at minimizing the ability of the ribosomes atoms to move around after their bonds are broken by the intense beam. By using special solvents mixtures, we developed a procedure, that enabled us to minimize the spread of damage caused by the radiation. That is, measuring the diffraction of small areas of cooled crystals, while minimizing the damage during measurements, and avoiding the spread of the broken atoms to all areas of the crystal that had not yet been exposed to the radiation. Using this method and some other experimental procedures developed years earlier at the Weizmann Institute of Science, we succeeded in maintaining the diffraction abilities of the ribosome’s crystals for a long time, and eventually—after 20 years—also deciphering its structure and understanding how it functions.
Throughout our entire project, many scientists thought it was impossible to decipher the structure of the ribosome. Well-known scientists even made fun of us for the substantial challenge we were trying to tackle. But thanks to our determination and creativity, we succeeded to accomplish our mission by constantly searching for “out of the box” solutions. In fact, despite the mistrust of our ability in the scientific world, and the wide contempt for our deep cooling procedure, the entire bio-crystallography adopted it within a few months. Our remarkable success also led to an important medical application, which you will read about in the next section.
Fighting for Health—How to Neutralize Bacteria
As mentioned earlier, the ribosome is a highly important component of every living cell. When the function of the ribosome is significantly impaired, the cell stops producing proteins and cannot continue to function. This is also true for bacteria. Bacterial ribosomes function similarly to human ribosomes but their structures are slightly, although significantly, different from that of human ribosomes. These differences allow us to use antibiotics that harm the bacteria without wounding the patients taking the antibiotics.
There are several antibiotic families. Natural antibiotics are produced by bacteria as weapons in the “wars” between different types of bacteria fighting for the same resources. They damage other bacteria by paralyzing an essential mechanism required for bacterial activity. One of these targets is the ribosome. Although the ribosome is huge compared to the size of the antibiotic, most antibiotics bind and block one of the active ribosomal sites: for example, they inactivate the PTC or block the NPET in the large ribosomal subunit, thus preventing the nascent protein chain growth (Figure 4) by discontinuing its elongation. Another ribosomal target is to block the movement of mRNA within the small ribosomal subunit. This blockade prevents the mRNA from being properly read, which is necessary for the production of a proper protein. Other antibiotics target the A site of the small ribosomal subunit and prevent the tRNA from binding to the ribosome. When this happens, the amino acid cannot bind to the next amino acid in the chain [4]. Click here to watch a video demonstrating how antibiotics can disrupt the function of bacterial ribosomes.
Indeed, the findings from our research have important applications in the field of medicine. But you may have heard that some disease-causing bacteria have developed resistance to certain antibiotics, making them even more dangerous. Understanding the structure and function of the ribosome has allowed us to better understand how different antibiotics work and how bacteria have developed resistance mechanisms to them, thus provided ideas of how to the fight against antibiotic resistance. This is an example of how the great perseverance and struggle we have invested over dozens of years in deciphering the structure and function of the ribosome have been justified and have borne fruit. In this context, I would like to share with you some recommendations that I have formulated as a result of this process.
Recommendation for Young Minds
My main recommendation is to follow your curiosity. If you try to contribute new scientific information to a scientific field you are extremely passionate about, you may face problems in searching for support—especially in the beginning. Some might say you have no chance of success. Others might say that your research is misleading, and they might even claim that you are wasting the public’s money. I recommend that you focus on your curiosity—be strong and believe in yourself even if you do not yet achieve the goal you wished for. If you delve deeper into the problem that interests you and its logical basis, you will increase the chances of its solution. My goal was to decipher the structure of the ribosome. Throughout the process, I believed that even if I did not succeed in deciphering the entire structure, I would make significant scientific progress along the way. Even before the structure of the ribosome was deciphered, my research led to the development of new methods in crystallography. These methods led to the solution of tens of thousands of previously unknown structures that helped improve our understanding of many biological processes.
Another aspect I would like to share with you is that the more challenging the path, the greater the satisfaction at its end. So be determined and work to overcome the difficulties that stand in your way, trusting that great satisfaction awaits you around the next corner.
Glossary
DNA: ↑ The genetic material passed from parents to their offspring. It contains the instructions to produce the cell components, called genes. DNA consists of four building blocks called nucleic acids.
Proteins: ↑ The “workers” of the cell—molecules that perform most actions in the cell, and in the body in general. These include, for example, producing movement (muscle activation), transporting oxygen and removing carbon dioxide from the blood (respiration), building the body’s skin and connective tissues (structure), and protecting the body from invaders (immune system).
Amino Acids: ↑ The building blocks of proteins. There are about 20 different amino acids. Within the proteins, the amino acids are linked together by a peptide bond. These bonds connect the amino acids into chains, and this is how the protein is formed. An average sized protein consists of 150–300 amino acids.
Peptide Bond: ↑ The chemical bond that links amino acids together in a protein.
Messenger RNA (mRNA): ↑ A type of RNA that copies the genetic information from DNA to the ribosome, where it serves as a template for protein synthesis. During the process of protein synthesis, the mRNA is read by the ribosome and used to assemble a chain of amino acids in the correct order according to the genetic code. The mRNA molecule produced by this process serves as an accessible and safe copy to be translated to proteins, and disintegrates once it is translated.
Transcription: ↑ The process by which a messenger RNA molecule is created based on the DNA molecule. DNA contains all the operating instructions for the production of proteins in the cell (the genes). Since DNA is the original copy and cannot be replaced, the cell has several different mechanisms to protect it, including the transcription mechanism.
Ribosome: ↑ The organelle in the cell that produces proteins. The ribosome consists of two subunits, the small ribosomal subunit, and the large ribosomal subunit. Both subunits are composed of many ribosomal proteins and ribosomal RNA. The small ribosomal subunit is responsible for decoding the genetic code according to the messenger RNA, while the large ribosomal subunit is responsible for protein production according to the instructions of the genetic material.
Translation: ↑ This is the second phase of protein synthesis, in which the genetic information encoded in messenger RNA is used to assemble a chain of amino acids by the ribosome. As the amino acids are added to the growing chain, the protein begins to advance within its tunnel toward exiting the ribosome and folding into its final three-dimensional structure.
Transfer RNA (tRNA): ↑ A family of unique RNA molecules each brings a specific amino acid to the ribosome during translation. Each tRNA is specific for a particular anticodon sequence that binds to a complementary codon on the mRNA and carries the amino acid decoded by the mRNA.
Ribosomal RNA (rRNA): ↑ A unique RNA sequence that forms the structural and catalytic core of the ribosome. About two-thirds of the mass of the ribosome is rRNA. The last third of its mass is ribosomal proteins.
Crystallography: ↑ A research technique used to determine the three-dimensional structure of molecules, including proteins and nucleic acids. It involves growing crystals of the molecule and radiating them with an X-ray beam to produce a diffraction pattern that can be used to calculate the positions of the atoms in the molecules within the unit cell. This technique is used to understand protein function and interactions.
Crystal: ↑ A solid material with a repeating pattern of atoms or molecules. This pattern gives crystals special properties, such as the ability to diffract light and generate electricity. Scientists use crystals to study the shape and internal bonds of molecules such as metals, organic compounds, proteins and DNA that are too small to see by a light microscope.
Conflict of Interest
NS declared that they were an employee of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Zehava Cohen for providing Figures 1, 2, 5.
References
[1] ↑ Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102:615–23. doi: 10.1016/S0092-8674(00)00084-2
[2] ↑ Zhou, X, Liao, W-J, Liao, J-M, Liao, P, Lu, H. 2015. Ribosomal proteins: functions beyond the ribosome. Mol. Cell. Biol. 7:92–104. doi: 10.1093/jmcb/mjv014
[3] ↑ Bashan, A, Agmon, I, Zarivach, R, Schluenzen, F, Harms, J, Berisio, R, et al. 2003. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell 11:91–102. doi: 10.1016/S1097-2765(03)00009-1
[4] ↑ Schlünzen, F., Zarivach, R., Harms, J., Bashan, A., Tocilj, A., Albrecht, R., et al. 2001 Structural basis for the interaction of antibiotics with the peptidyl transferase center in eubacteria. Nature 413:814–21. doi: 10.1038/35101544
