Abstract
In this article, we provide a brief overview of network science by highlighting the importance of network models. We will discuss the origins of networks and describe early studies. Then we will explain the important role of gene networks in biology for understanding the way genes cause certain physical traits in organisms. Aside from biology, networks can be found in essentially all areas of science, including chemistry, medicine, business, finance, and the social sciences. Our digital society generates large amounts of data and networks can be created from these data by using statistical methods. Those networks can then be used to help us understand various aspects of society and to ultimately improve our lives.
What Are Networks And Where Do They Come From?
The Tower of Babel tells the biblical story that God confused the language of men trying to build a tower reaching heaven, by introducing multiple languages among them. In this way, the men could no longer understand each other and had to give up. In a sense, networks provide a mathematical language that allows scientist from many different fields to understand each other. This makes networks important tools that allow us to work on the most difficult problems imaginable [1]. The field for using networks to solve complex problems is called network science [2].
The terms graph and network are often used interchangeably. Traditionally, a graph refers to a mathematical object that does not need to have a real-world representation. Although mathematicians have studied networks for over 200 years (by people like Euler and Cayley), the idea of a graph is much more recent and can be traced back to the mathematician König in the 1930s < 100 years ago.
In its most simple form, a graph is mathematically defined by the following definition:
Definition 1.1: The pair G = (V, E) where V represents a finite set of vertices and E the set of edges, is called a finite undirected graph.
This definition can be understood in the following way: a graph is made of just two entities, nodes also called vertices (represented by V) and edges also called links (represented by E). Here V is a set containing some elements, e.g., V = {a, b, c, …} and E is another set containing information about the connections between the elements in V, e.g., E = {(a, b), (a, c), …}. The term denotes another set which contains all possible pairs of nodes one can form from the nodes contained in the set V. Hence, the meaning is similar to binomial coefficients if V would be a positive integer number. The symbol ⊆ in means that all elements in the set E are also contained in the set and, hence, E is a subset of . This includes the case where all elements in are also in E. In Figure 1 (left), we show a concrete example for a network.
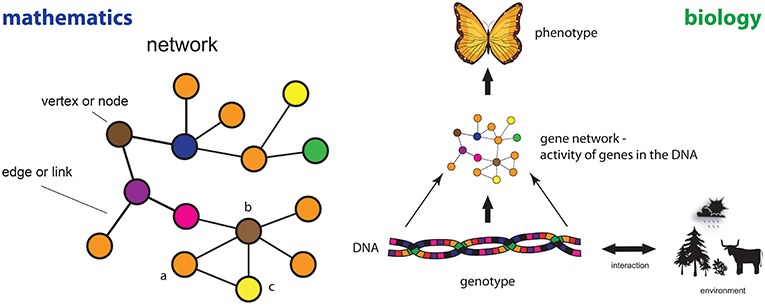
- Figure 1 - Left: An example of a mathematical network and the parts that define it.
- Right: The principle of the epigenetic landscape showing a gene network as the connection between the genotype of an organism (its DNA) and its phenotype (the way the organism looks).
The reader may wonder about the way the mathematical definition of the network is written. This is what is called abstract. If one wants to study networks on a serious level such formulations are inherent to the field and a keen interest in abstractness in general is required in one’s skill set.
Among the first mathematical networks that have been studied are random networks. Erdös and Rényi studied these in the 1960s. A random graph with N nodes is obtained by connecting every pair of vertices with a fixed probability p. Despite its simple construction, studying its mathematical properties is surprisingly complex and generations of scientists have worked on this problem.
Where Do We Find Networks?
In biology, the importance of networks has been recognized because biological processes and systems need to be studied holistically (concerning every part) [3]. That means biological systems cannot be reduced to arbitrarily small parts, but the minimal size of such a part still needs to be functional in a sense that the underlying organisms work.
One of the first insights in this respect is from Conrad Waddington, who conceived the idea of the epigenetic landscape in the 1940s [4]. Here epigenetic means the study of heritable phenotype (see Figure 1) changes that do not change the DNA. Its basic idea is shown in Figure 1 (right). On a molecular level within a biological cell of an organism (plant, animal, or human), the interactions between genes and gene products (proteins) can be represented as a gene network, e.g., as a transcriptional regulatory network or a protein network. In this network nodes correspond to genes and edges correspond to interactions between genes. This means that networks appear naturally in studying molecular interactions as their graphical visualization and mathematical representation [5]. Depending on the content of the DNA of an organism and its interaction with the environment (diet, life style etc.) the structure of the gene network changes because the activity patterns of the genes change and as a consequence the phenotype (physical appearance, e.g., color and shape of wings of a butterfly) of an organism emerges.
Networks Can Be Used In Many Different Fields
Networks are very flexible tools and they can be used in many fields besides biology. This flexibility is due to three major characteristics of networks. First, networks can make the complex interactions between all the different parts of a system visible. Second, networks form a mathematical representation of the system that can be studied and manipulated. Third, a network represents a data structure that can be conveniently stored on a computer and analyzed in many different ways, for example, using statistical methods for finding tightly connected communities of nodes.
Because of this flexibility, networks are used in many different fields, including chemistry, physics, biology, medicine, business, finance, and social media. Here is a list of the most important networks from these fields and what they can be used to study.
- Chemical structures, to study the way chemical compounds are related to each other.
- Metabolic networks, to study how organisms do things like digest food, grow, and develop.
- Signaling networks, to study the molecular communication between proteins.
- Transcriptional regulatory networks, to study the activation of genes.
- Protein interaction networks, to study complex formation.
- Financial networks, to study optimal portfolios.
- Graph-based document structures, to study writing styles.
- Consumer behavior networks, to study online shopping habits.
- Economic networks, to study global trading patterns.
- Social networks, to study human relations.
In Figure 2, you can see two examples of real networks. These networks are a special type called a bipartite network. Bipartite networks consist of two type of nodes, shown by the two colors (blue and orange). The meaning of these nodes depends on the problem being studied. For the social network shown, the blue nodes correspond to actors and the orange nodes to movies in which the actors played. For instance, the fact that Harrison Ford played in Star Wars is represented in Figure 2. For the financial network shown, the blue nodes correspond to investors and the orange nodes to stocks bought by the investors. We showed the social network and the financial network overlain by each other because we wanted to again emphasize the abstract character of networks. With respect to these two networks, you can practice making the situation concrete by focusing on one network at a time and ignoring the other one completely. This exercise teaches you how to deal with this abstract situation.
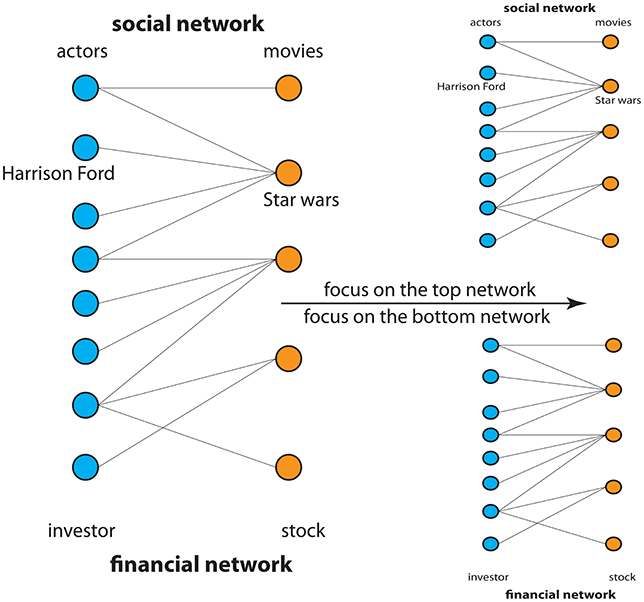
- Figure 2 - Shown are two examples for the application of networks representing a social network and financial network.
- To make the network on the left concrete focus, first, on the top network and then on the bottom network only. This gives two individual networks. This clarifies the abstract meaning of the left network.
What Do Networks Mean?
The concrete meaning and the usage of networks depends on the problems they represent. For instance, we might use a social network to look at actors connected to the same movies, to see which actors appeared in the same movies together. Practically this could correspond to a grouping of actors according to movie genres. For a graph model of a protein one might study modules or community structures of vertices—practically these could correspond to evolutionary conserved domains of proteins. For a computer network one could study how many vertices can be removed from the network but still keep it connected—practically this could indicate the robustness of such a network with respect to hacker attacks. For a transcriptional regulatory network one could identify the nodes having the highest number of connections to other nodes—practically this defines hub genes and could indicate the importance of such genes.
These are just a few examples describing how abstract networks can be used to solve concrete problems in various fields of research.
How Do We Get the Networks?
In contrast to the networks shown in Figure 2, which are fairly simple and easy to understand, there are more abstract networks. Such networks need to be inferred from data by applying methods from statistics and machine learning. In Figure 3, we give an overview of aspects of our digital society that allows us to generate massive amounts of data about almost every aspect of life, including health (medical tests, smartphones, smartwatches etc.), business (stock market, Amazon etc.) and social media (Twitter, Facebook etc.). Data from these areas can be analyzed with the help of computers to produce network models. These network models can provide us with novel insights about many aspects of our society, such as the economy, methods of patient care, or consumer behavior, that can then be used to help us to improve our lives.
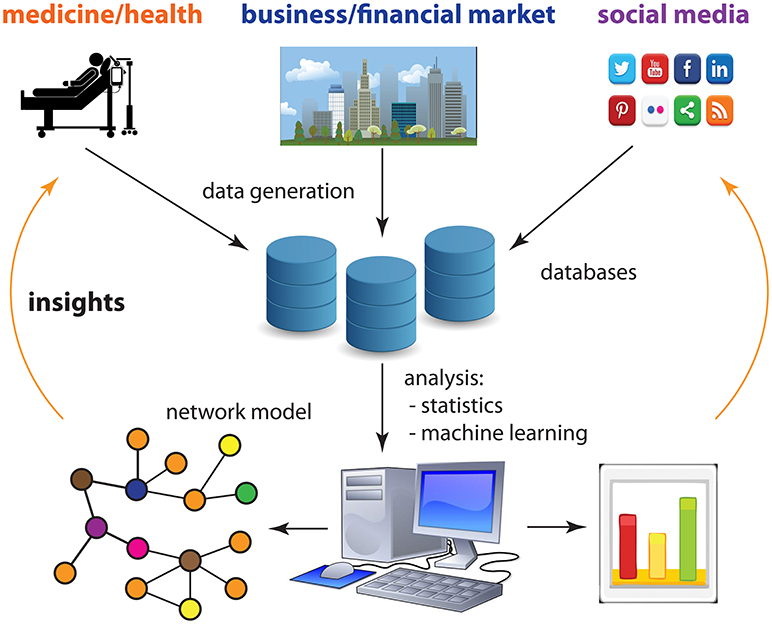
- Figure 3 - Our digital society allows us to generate massive amounts of data about almost every aspect of life, including health, business, and social media.
- These data can be analyzed with computers to create network models that teach us new things about our society and allow us to make improvements in those areas of society.
Conclusion
We hope that our brief overview showed that networks provide a fundamental language that allows us to tackle the most interesting and important problems in society and science. However, to be able to study such network models, strong understanding of mathematics is required.
Author Contributions
FE-S conceived the study. All authors contributed to all aspect of the preparation and writing of the paper. All authors approved the final version.
Glossary
Mathematical Definition: ↑ A description expressed in the language of mathematics that is very important.
Epigenetic: ↑ Is the study of heritable phenotype changes that do not involve alterations of the DNA. That means components that are on top (the greek prefix epi means ‘on top’) of the genes.
DNA: ↑ Deoxyribonucleic acid, a self-replicating material which is present in nearly all living organisms as the main component of chromosomes, and carrier of genetic information.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
[1] ↑ Emmert-Streib, F., Dehmer, M., and Shi, Y. 2016. Fifty years of graph matching, network alignment and network comparison. Inform. Sci. 346–347:180–97. doi: 10.1016/j.ins.2016.01.074
[2] ↑ Barabási, A.-L. 2013. Network science. Philos. Trans. R. Soc. A 371:20120375. doi: 10.1098/rsta.2012.0375
[3] ↑ Emmert-Streib, F., and Dehmer, M. 2011. Networks for systems biology: conceptual connection of data and function. IET Syst. Biol. 5:185. doi: 10.1049/iet-syb.2010.0025
[4] ↑ Waddington, C. H. 1957. The Strategy of the Genes. London: George Allen & Unwin.
[5] ↑ Emmert-Streib, F., and Glazko, G. V. 2011. Network biology: a direct approach to study biological function. Wiley Interdiscipl. Rev. Syst. Biol. Med. 3:379–91. doi: 10.1002/wsbm.134