Abstract
Each of us has a mix of microbes that colonizes our intestines, as unique as our fingerprints. Most microbes are beneficial and are essential for keeping us healthy. If the microbes in our intestines are out of balance and too many harmful microbes take control, intestinal diseases might result. Recent advances in computers and laboratory methods have helped scientists understand much more about our intestinal microbes and their unique signatures: who they are, what they do, and which useful or harmful substances they produce. We wanted to know which bacteria can cause intestinal diseases, and whether we could use these bacterial signatures to foresee the futures of patients. We analyzed the bacteria in patients with intestinal diseases and identified certain bacteria that can produce harmful compounds. This information could help doctors predict who is more at risk of developing intestinal disease and which treatment could work best for each patient.
Your Microbes—Your Identity
Microbes are tiny creatures that are impossible to see without a microscope. Trillions of microbes live in and on our bodies, especially in our intestines. The gut microbiota is the term we use to describe this entire vast and diverse collection of bacteria and other organisms that live in our intestines. Unfortunately, we mostly associate bacteria with illness. However, most of the bacteria in our bodies are beneficial to human health. They help us break down and digest the foods we eat, produce essential vitamins, and fight off harmful bacteria. In return, we offer them a place to live and the nutrients they need to thrive. Beneficial bacteria are also important to create a powerful immune system. The immune system protects our bodies against illnesses. When the number of harmful bacteria is kept to a minimum, they generally do not cause problems. But under certain conditions, harmful bacteria can multiply and kick out beneficial bacteria, which can result in multiple diseases [1].
Is everyone’s gut microbiota the same? Yes and no! While one-third of the gut microbiota is common between individuals, two-thirds of each person’s microbiota is unique to them and as personal as their fingerprints! While everyone’s gut microbiota performs similar functions, these functions are not necessarily carried out by the same bacteria in everyone’s body [2]. You could think of the intestines as a city for microbes. Every city has its own firefighters, police officers, bus drivers and garbage collectors. Different people perform these roles in each city and within each city, those people can change over time. In the bacterial world, hundreds of microorganisms can perform essential roles such as producing vitamins or breaking down the carbohydrates in the foods we eat [3]. This means that, even if our bacterial communities look different, they are doing the same set of important jobs. In a city, if many productive residents live there and if they can perform multiple essential jobs, the city will flourish. Similarly, we are healthier if our intestines are home to a high number of beneficial bacteria that can perform a variety of functions (Figure 1).
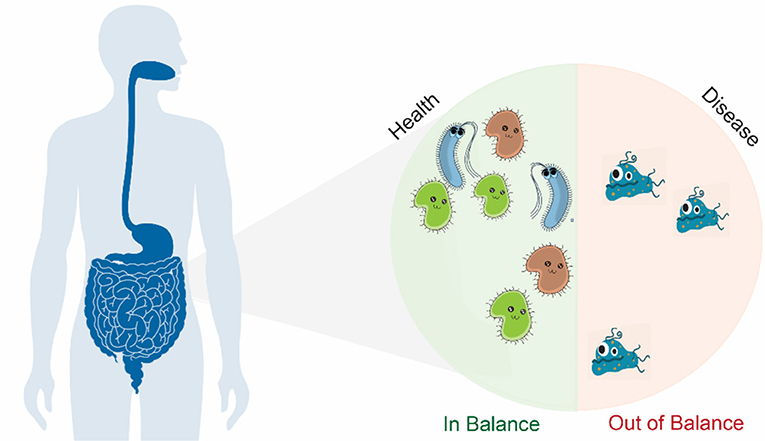
- Figure 1 - Trillions of bacteria live in and on our bodies.
- Most of them live in our intestines. To stay healthy, we need a wide range of bacteria that can perform many functions essential for our health. It is important for the number of beneficial bacteria to remain higher than the number of harmful ones. In patients with intestinal diseases, fewer bacteria live in their intestines and harmful bacteria take over and produce toxic substances that lead to disease.
The Connection Between Gut Bacteria and Crohn’s Disease
Scientists have been studying whether changes to our intestinal bacteria are somehow linked to the foods we eat, the medications we take, or to whether we are healthy or sick. We were particularly interested in understanding whether the bacteria that live inside people who have long-lasting intestinal inflammation (also called Crohn’s disease) have unique and harmful characteristics. Crohn’s disease was named after the doctor who first described it in 1932, and it affects millions of people worldwide. Crohn’s disease affects teens and young adults, but mostly people 20–40 years of age. In Crohn’s disease, parts of the intestine become red and swollen. Patients can have times when they get very sick (what we call flare-ups) and other periods when they feel completely fine (what we call remission). Patients with Crohn’s disease suffer from bloody diarrhea, abdominal pain, weight loss, fever, and loss of appetite.
Normally, the immune system watches out for danger. However, in Crohn’s disease, the immune system gets confused and attacks both the good and the bad bacteria. Disturbing this bacterial balance increases the toxins produced by harmful bacteria and causes destruction of the intestinal tissue. Medicines can help reduce the immune system attacks, but intestinal tissue damage gets worse over time. Research showed that patients with Crohn’s disease have bacterial communities that look quite different from those found in healthy individuals. These bacterial communities were mostly dominated by bacteria with the potential to become harmful under certain conditions.
Who Is There? What Are They Doing?
To figure out which bacteria are causing Crohn’s disease, we asked patients to collect stool samples (poop) during both remission and flare-ups. We performed laboratory experiments to measure the molecules and compounds present in the poop samples. We also broke open the bacterial cells to release the DNA so we could analyze the bacterial genes and identify which bacteria were present. Finally, we used computers and statistics to understand the relationships between the bacteria, their products, and the health status of patients. All of this information gives us specific patterns, called bacterial signatures [4, 5], for remission and flare-ups (Figure 2).
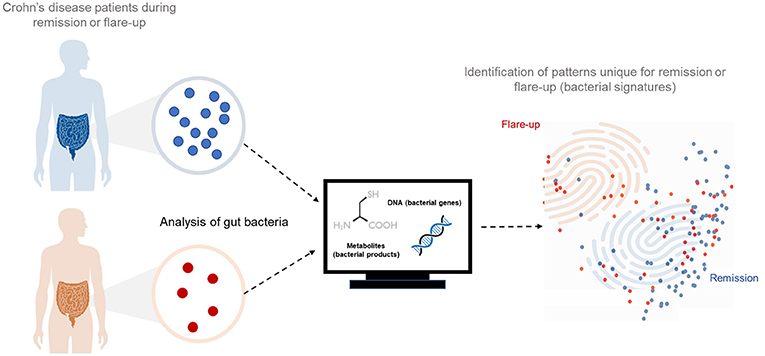
- Figure 2 - We took samples of intestinal bacteria from Crohn’s disease patients who were either in remission or experiencing flare-ups.
- We did experiments to determine which metabolites were being made by the bacteria and we broke open the bacterial cells to study their genes. We analyzed all the data using computers and mathematics. In the end, we could see unique patterns, which we called bacterial signatures, for each condition.
We found a big difference in the numbers of microbes in the intestines of patients during periods of remission vs. flare-ups. Many more bacteria were present during remission, and they were also much more diverse, meaning there were more types of bacteria present. In addition, we saw big changes in the numbers of specific types of microbes between these two groups. For example, there were more bacteria from the groups Escherichia coli and Desulfovibrio during flare-ups. To learn more about these bacteria, we measured the concentration of the metabolites they produce. Metabolites are small molecules that take part in, or are produced by, the chemical reactions of metabolism. The concentrations of certain metabolites in poop, urine, or blood are linked to the risk of developing certain diseases. When we compared patient samples from remission vs. flare-ups, we found differing amounts of certain metabolites.
Teaching Computers to Tell the Future
Next, we wondered if we could predict whether patients with Crohn’s disease would remain in remission or have flare-ups, based on their bacterial signatures. To do so, we used machine learning, in which computers are trained to perform complex tasks. For a computer to identify the bacterial signature of a patient who will soon get a flare-up, the computer is given many examples of bacterial signatures from patients during flare-ups, so that the computer learns the patterns of what a flare-up bacterial signature looks like. This phase is called learning. Then, we test the computer on what it learnt by challenging it with examples of bacterial signatures where the computer needs to decide whether each signature came from a healthy subject or a patient having a flare-up. This phase is called testing. If the model passes our tests, we can rely on it to tell whether a patient is healthy or will soon have a flare-up from the bacterial signature. This phase is called prediction. To our excitement, our computer was correct in its prediction of flare-ups 96% of the time (Figure 3)!
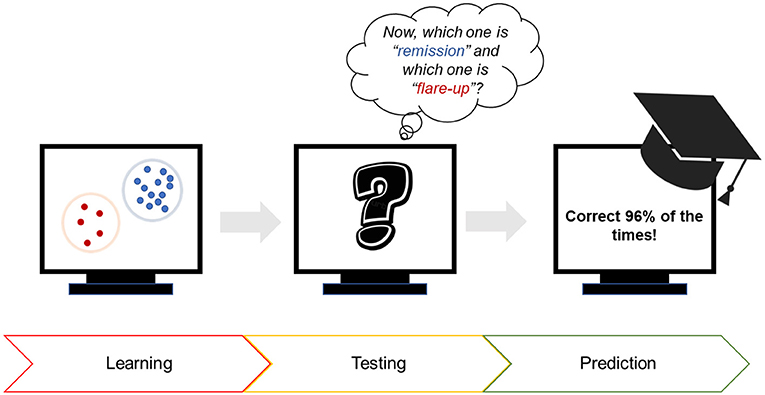
- Figure 3 - Using computers to predict the occurrence of flare-ups in Crohn’s disease patients.
- First, the computer was shown many bacterial signatures of patients during flare-ups. Then, the computer was tested to see if it could identify known signatures as “remission” or “flare-up.” Third, we used the computer to predict whether a patient was about to experience a flare-up. The computer was correct 96% of the time!
Microbial Treatments for Intestinal Disease
Our research could be useful for treating patients with intestinal diseases like Crohn’s disease. If we can use machine learning to analyse bacterial signatures in patients’ poop, this could help us tell whether the patient is healthy or might develop intestinal disease in the future. If we can identify the exact types of bacteria that are decreasing when a patient has a flare-up, we might be able to collect those bacteria, let them grow and multiply in the lab, and make them into a pill that we can give to patients to help them improve. Such beneficial bacteria are called probiotics. We all have some probiotic bacteria in our intestines, but they are also found in foods including yogurt, cheese, bread, or buttermilk.
If healthy people have enough beneficial microbes, they can donate poop samples that we could then transfer into the colons of patients with poor bacterial signatures. This is called fecal microbiota transplantation. Transplanting the healthy microbes into patients could kick-start the bacterial community and restore balance to the gut microbes. There is still some work to do before we can effectively treat patients using the information from our study. The work we have done so far is already important because the more we know about gut bacteria and their functions, the more likely we will be to find new, effective treatment options for the millions patients across the globe who suffer from intestinal diseases like Crohn’s disease.
Glossary
Gut Microbiota: ↑ The group of microorganisms that live in human intestines. Most of the organisms are beneficial and perform functions that keep us healthy.
Flare-ups: ↑ Periods of time when a patient with a long-lasting disease shows more symptoms and feels worse.
Remission: ↑ Periods of time when a patient with a long-lasting disease shows no symptoms and feels better.
Bacterial Signature: ↑ Unique patterns of bacterial community composition and function that can differentiate between physiological and pathological conditions and can be used to predict the risk of disease development or progression.
Metabolites: ↑ Small chemical compounds that are involved in or produced by an organism’s metabolism. In this research, we looked at metabolites produced by bacteria.
Machine Learning: ↑ A branch of science which allows computers to perform tasks without us having to tell the computer how to do it. The computer learns to perform a task by observing a lot of examples and trying to figure out how to make the best judgement.
Probiotics: ↑ A group of beneficial bacteria that people can consume to restore balance to their gut bacteria.
Fecal Microbiota Transplantation: ↑ A procedure in which poop from a healthy donor is transplanted in the colon of a patient. The good bacteria replace the bad ones, and a healthy balance is restored.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Leona and Harry Helmsley Charitable Trust (IBDOT Consortium). The authors would like to thank the entire research team who joined for this amazing discovery journey exploring microbes. Special thanks to Dr. Eva Rath for proof-reading this article. Finally, Amira would like to thank her daughter Laila for her help with the figures and for her curiosity while discussing and reviewing this article.
Original Source Article
↑Metwaly, A., Dunkel, A., Waldschmitt, N., Raj, A. C. D., Lagkouvardos, I., Corraliza, A. M., et al. 2020. Integrated microbiota and metabolite profiles link Crohn’s disease to sulfur metabolism. Nat. Commun. 11:4322. doi: 10.1038/s41467-020-17956-1
References
[1] ↑ Buttó, L. F., and Haller, D. 2016. Dysbiosis in intestinal inflammation: cause or consequence. Int. J. Med. Microbiol. 306:302–9. doi: 10.1016/j.ijmm.2016.02.010
[2] ↑ Consortium, H. 2013. Structure, function and diversity of the healthy human microbiome. Nature. 486:207–14. doi: 10.1038/nature11234
[3] ↑ Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature. 489:220–30. doi: 10.1038/nature11550
[4] ↑ Metwaly, A., Reitmeier, S., and Haller, D. 2022. Microbiome risk profiles as biomarkers for inflammatory and metabolic disorders. Nat. Rev. Gastroenterol. Hepatol. 19:383–97. doi: 10.1038/s41575-022-00581-2
[5] ↑ Waldschmitt, N., Metwaly, A., Fischer, S., and Haller, D. 2018. Microbial signatures as a predictive tool in IBD—pearls and pitfalls. Inflamm. Bowel Dis. 24:1123–32. doi: 10.1093/ibd/izy059

