Abstract
There are many organisms living in lakes, for example, fish, aquatic plants, microalgae, and bacteria. But have you wondered what organisms have inhabited a lake throughout its history? Are there any species that are no longer found in the lake today? Has the ecosystem changed over time? When they die, most lake organisms leave their remains (pollen, shells, fossils, and DNA). Remains are preserved for many years in the sediments deposited at the lake bottom. Scientists are using an exciting technology that identifies organisms from DNA extracted from sediments that are over 100 years old. In this article, we will tell you how DNA is preserved in sediments at the bottom of lakes and how it is used to find out which organisms were present in the past and which are still living in a lake today.
Lakes and Their Biodiversity
Lakes are bodies of freshwater formed in hollows at the Earth’s surface. There are different types of lakes depending on how they form. Glacial lakes are formed by melting ice. Volcanic lakes store water in inactive craters. Artificial lakes are created by humans who excavate the ground and fill the area with water.
Lakes are important because they store fresh water, which can be used for agriculture, energy generation, recreation, and as drinking water.
Lakes provide a home to a wide variety of living things. In lakes, we can find many kinds of fish, water birds such as ducks and herons, amphibians such as frogs, reptiles, aquatic plants, and insects. A lake’s biodiversity can change over time due to changes in lake conditions.
The biodiversity of a lake is related to the region’s climate. Changes in temperature, rainfall, or water quality can change lake’s biodiversity.
For example, the species that inhabit a lake are adapted to specific water temperatures. When the water temperature changes drastically, species can die or migrate. When a lake receives less rain, biodiversity may change because the lake dries out and water-dependent species can die. Changes in water quality, such as pollution, can also affect the types of organisms that live in a lake. When the water is more contaminated, it causes health problems for the plants and animals that live there.
When lake organisms die, they are buried in sediments at the bottom of lakes. Some organisms have hard parts, like bones or shells, that can fossilize and be preserved in rock or sediment for a long time. Examples range from a huge dinosaur to microscopic algae called diatoms (see this Frontiers for Young Minds article). But some organisms have only soft parts, and they break down in sediments over time. Scientists can identify fossilized organisms preserved in sediments by eye if they are large, and using a microscope if they are small.
However, to study organisms that do not fossilize, scientists must use specialized techniques. One of them is DNA analysis, which is used to identify the type of living beings through the study of the genes. For example, scientists discovered the diversity of organisms that have inhabited Lake Chalco (Mexico) throughout its history using DNA analysis [1].
What are Lacustrine Sediments?
Sediments are mainly formed by the breakdown of rocks over long periods of time. The sediments that deposit at the bottom of a lake are specifically called lacustrine sediments. They originate from external materials that enter the lake through rivers or through the air, like soil and animal waste, and from materials within the lake, including shells, fossil remains, DNA, and proteins [2].
Each season, a new sediment layer is deposited over the sediment layer from the previous year. Thus, the deeper the sediment, the older it is! This means that lacustrine sediments can provide evidence of a lake’s history.
Scientists use sediments to find out what a lake was like in the past and to understand how species have changed through time. Can you imagine knowing what kinds of fish, microbes, or insects lived in a lake 5, 50, or 100 years ago? Scientists can use DNA analysis to learn which species inhabited a lake in the past, even if those organisms did not leave fossil remains [3, 4].
Obtaining Lacustrine Sediments From a Lake
To perform DNA analysis, researchers must first extract lacustrine sediments from a lake. From boats, researchers can use various methods to recover sediments from the lake bottom. For example, they can use an instrument called a hand corer, which they drop from the boat to the lake bottom, where it penetrates the lacustrine sediments (Figure 1). The sample that is collected is stored in a transparent plastic tube. The tube is then opened in the lab so that scientists can describe the sediments and take smaller samples for other analyses. It is important for scientists to wear lab coats, masks, and gloves to avoid contaminating the samples.
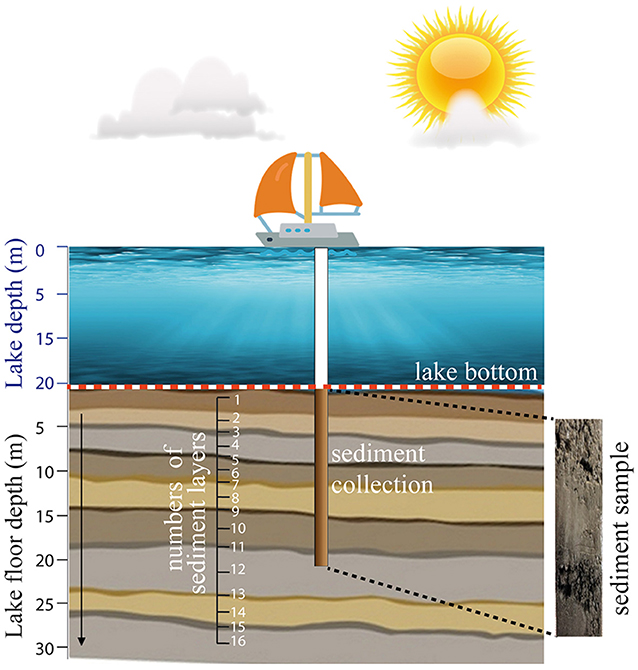
- Figure 1 - Sediment extraction from a lake using a device called a hand corer.
- The device is lowered from a boat and it penetrates the sediment layers to remove a cylinder of sediment, which is stored in a transparent plastic tube and then studied in the lab. On the right, you can see an example of a sediment sample with several layers visible.
Beyond Fossil Remains
DNA is found in the cells of all living organisms. It contains the necessary information for building the organism’s body and maintaining its life processes. DNA is made from four compounds called nitrogenous bases: adenine, guanine, thymine, and cytosine (A, G, T, and C). The order of these compounds, called the DNA sequence, varies from organism to organism almost like a barcode that can be used to identify organisms. When lake organisms die and are buried in the sediments, the DNA can persist in the sediments for many years. This means that sediments can store information about the organisms that currently live in a lake or that have lived there in the past [5].
In recent years, scientists have used DNA analysis to reconstruct the history of lakes and their organisms. DNA analysis can detect all kinds of organisms, including plants, animals, and even endangered species.
How is DNA Preserved in Lacustrine Sediments?
Certain qualities of the lake bottom help to preserve DNA for many years, for example, low light, low oxygen, and cold temperatures. Scientists still do not know exactly how long DNA can survive in lake sediments, but some studies have shown that DNA in lacustrine sediments is up to 11,000 years old [6]!
But how exactly is DNA preserved in lacustrine sediments, and how is it protected from being broken down? DNA in lacustrine sediments can either be inside a cell (from an organism that recently died) or outside a cell. When a cell breaks down, it releases its DNA into the sediments. Often, substances called nucleases, which are present in the sediments, can act like Pac-Man and break down the DNA. In this case, DNA is no longer available for scientists to study. But DNA can also stick to organic matter, minerals, and clays in the sediments, which protects DNA against nucleases and preserves it. DNA can also be preserved when it is taken up from the environment by certain other cells, which store the DNA fragments along with their own DNA (Figure 2) [7]. This preserves the DNA as well.
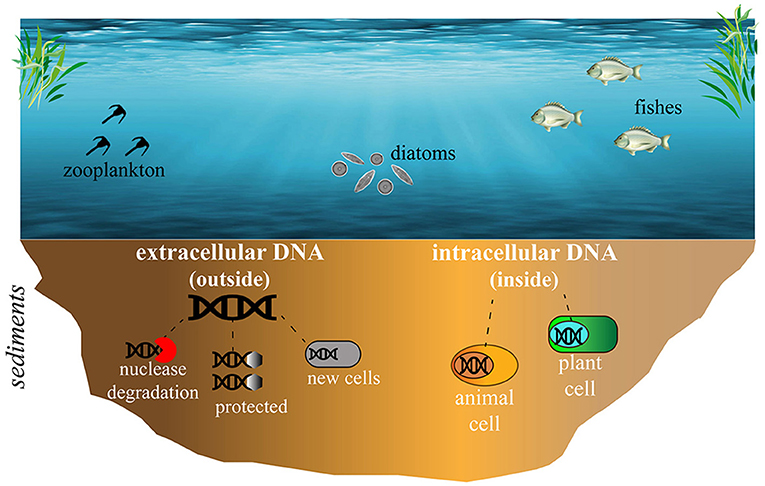
- Figure 2 - In lake sediments, DNA can be found either outside of cells or inside of cells.
- When animals and plants die, DNA can be found inside their cells for a while, until those cells break down. Once it is outside of cells, DNA is sensitive to breakdown by substances called nucleases. However, if it sticks to clay, minerals, or organic matter in the sediment, DNA can be protected from nucleases. DNA can also be protected from nucleases if it is taken up by other cells and stored alongside their own DNA.
Who Does the DNA Belong To?
To find out which organisms have lived in a lake, scientists first isolate the DNA fragments from their sediment samples. Since there might be only tiny amounts of DNA in the samples, the DNA fragments are multiplied using a laboratory technique called PCR amplification. This works like a DNA copy machine to provide billions of copies from a single fragment, making the DNA much easier to study. The sequences of nitrogenous bases in the DNA fragments are then “read” with specific machines that show the order of the bases. Finally, when scientists have the sequences, they use a technique called bioinformatics to compare the DNA sequences with known sequences in big databases. This allows them to identify which organisms the DNA came from (Figure 3).
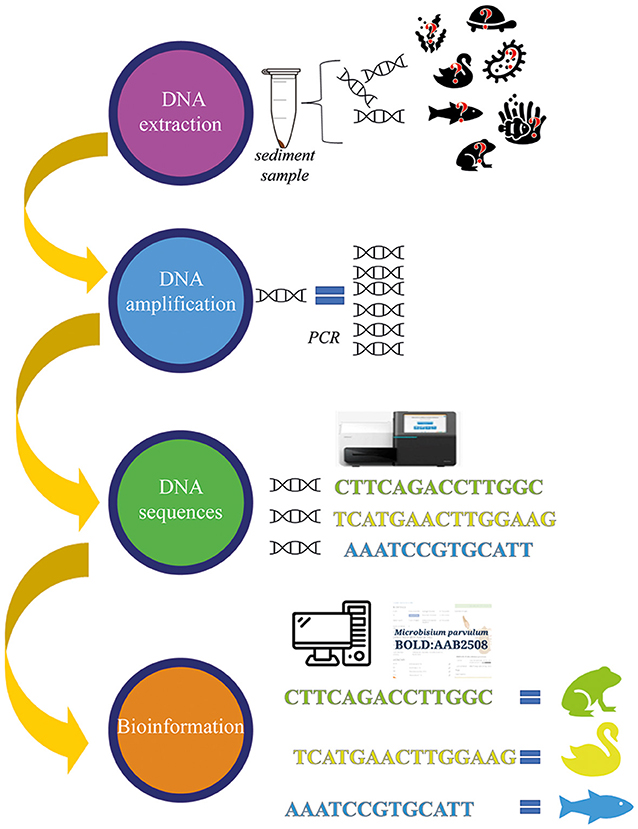
- Figure 3 - To identify organisms with DNA analysis, the DNA must first be isolated (extracted) from the sediment samples.
- Then, the DNA fragments are copied many times by a technique called PCR amplification. Once researchers have many copies of the DNA, they can use equipment to “read” the sequence of bases. These sequences can be compared to a database of known sequences to identify which organisms the DNA came from.
Exploring the Incredible History of Lakes
Sampling and analyzing the DNA preserved over many years in sediments at the bottom of lakes is interesting and exciting. DNA contains information about the lake’s history and can teach us how ecosystems have changed over time. Historical data are important for detecting which species are common within a lake, as well as those that are rare and/or those that do not fossilize. DNA analysis is an incredible scientific technique! However, more research is needed to understand exactly how many years DNA can be preserved in sediments and how it can change over time. Information about a lake’s history can help us to propose solutions for protecting and keeping lakes healthy.
Glossary
Biodiversity: ↑ Living beings that exist on the planet.
Fossilize: ↑ Processes that allow an organism to be preserved over time (e.g., a bone, a tooth).
Genes: ↑ A part of DNA that determines a characteristic of a living being.
Nitrogenous Bases: ↑ Compounds that make up the unique sequence of the DNA molecule. There are four bases, abbreviated A, C, T, and G.
Nucleases: ↑ Substances that break down DNA.
PCR Amplification: ↑ A technique that allows scientists to make millions of copies of a single DNA fragment, making it easier to study.
Bioinformatics: ↑ Using computers and software to make sense of complex biological data, like DNA sequences.
Acknowledgments
GAZ and MCDR-C are members of the National Council of Humanities, Science and Technology (CONAHCyT), Mexico, whereas ELP is a member of the National Scientific and Technical Research Council (CONICET), Argentina. CSM-R was supported by a fellowship from CONAHCyT. The authors want to thank Adriana Naranjo Vallejo and Ricardo Naranjo Vallejo for their contribution in revising the English version of this paper. Special thanks to young reviewers and editors for their suggestions that helped to improve the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
[1] ↑ Moguel, B., Pérez, L., Alcaraz, L. D., Blaz, J., Caballero, M., Muñoz-Velasco, I., et al. 2021. Holocene life and microbiome profiling in ancient tropical Lake Chalco, Mexico. Sci. Rep. 11:13848. doi: 10.1038/s41598-021-92981-8
[2] ↑ Schnurrenberger, D., Russell, J., and Kelts, K. 2003. Classification of lacustrine sediments based on sedimentary components. J. Paleolimnol. 29:141–54. doi: 10.1023/A:1023270324800
[3] ↑ Capo, E., Domaizon, I., Maier, D., Debroas, D., and Bigler, C. 2017. To what extent is the DNA of microbial eukaryotes modified during burying into lake sediments? A repeat-coring approach on annually laminated sediments. J. Paleolimnol. 58, 479–495. doi: 10.1007/s10933-017-0005-9
[4] ↑ Capo, E., Monchamp, M. E., Coolen, M. J., Domaizon, I., Armbrecht, L., and Bertilsson, S. 2022. Environmental paleomicrobiology: using DNA preserved in aquatic sediments to its full potential. Environ. Microbiol. 24:2201–9. doi: 10.1111/1462-2920.15913
[5] ↑ Domaizon, I., Winegardner, A., Capo, E., Gauthier, J., and Gregory-Eaves, I. 2017. DNA-based methods in paleolimnology: new opportunities for investigating long-term dynamics of lacustrine biodiversity. J. Paleolimnol. 58, 1–21. doi: 10.1007/s10933-017-9958-y
[6] ↑ Parducci, L., Bennett, K. D., Ficetola, G. F., Alsos, I. G., Suyama, Y., Wood, J. R., et al. 2017. Ancient plant DNA in lake sediments. New Phytol. 214, 924–942. doi: 10.1111/nph.14470
[7] ↑ Pedersen, M. W., Overballe-Petersen, S., Ermini, L., Sarkissian, C. D., Haile, J., Hellstrom, M., et al. 2015. Ancient and modern environmental DNA. Philos. Transact. R. Soc. B Biol. Sci. 370:20130383. doi: 10.1098/rstb.2013.0383
