Abstract
Lately, there have been more foodborne “superbug” outbreaks than ever before, which creates a problem because superbugs are antibiotic-resistant bacteria that are difficult to treat. To reduce such outbreaks, better ways of finding the source of the infection are needed. Superbugs, such as Salmonella are often transmitted through food. The world’s food supply system has become so complex that it is often difficult to find the source of an outbreak with older testing methods. A new method called whole genome sequencing (WGS) has now been developed to track superbug infections. Using WGS, it is now possible to identify the source of an outbreak in one country that may be transmitted through food imported from the opposite side of the world. Good outbreak tracing methods help scientists make better predictions about outbreaks. Finding the source of an outbreak early on can lead to better containment and lower costs.
What Are Superbugs?
Have you heard the term “superbug” and if so, do you understand what it means? Well, a superbug is simply a bacterium that causes sickness and is not easily treatable using antibiotics. Bacteria become superbugs by gaining genes that make them immune to antibiotics. A bacterium typically obtains superbug status when it is no longer able to be killed by two or more antibiotics. Usually, the development of a superbug is a slow process, but these days, people are using antibiotics when there is not a real need to use them. Excessive or incorrect use of antibiotics speeds up the evolution of superbugs. With no good way to treat superbugs, they have turned into a big problem, infecting thousands of people. If superbugs continue to evolve at the current rate, the world population could be hugely affected: by the year 2050, more than 400 million people could die from superbug infections [1].
How Do Foodborne Outbreaks Happen and What Are We Doing About It?
The World Health Organization has identified 12 bacteria that are superbugs. Among these, a bacterium named Salmonella is particularly problematic because it can not only infect animals but can survive in soil, water, and food. There are more than 2,000 different kinds of Salmonella [2]. Among these, only about 50 types cause human disease and have superbug status. The types of Salmonella that cause human infections are passed on to people through direct contact with Salmonella-colonized animals or when people eat contaminated food (Figure 1).
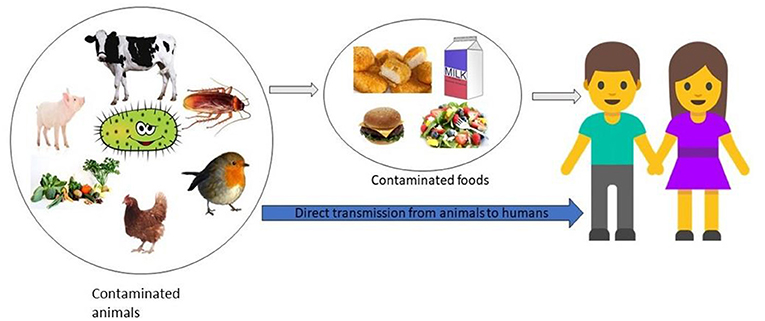
- Figure 1 - How do people get infected with superbugs?
- Superbugs, like some types of Salmonella, can live on animals, birds, insects, and vegetables. When people come in direct contact with Salmonella-colonized animals or when they eat contaminated food, they can become infected.
Contaminated food is a common source of Salmonella outbreaks. Food can be contaminated at multiple different stages in the supply chain. For example a chicken could catch Salmonella at the farm or its meat me be exposed to Salmonella at the meat packaging facility. In a foodborne Salmonella outbreak, a large number of people are often infected in a short time. This happens because the world’s food supply system is very complex. For example, in the United States, food comes from many national or international suppliers who then package and distribute food through a complicated supply system (Figure 2). Contamination anywhere in the supply system could expose millions of people to infection, which triggers an outbreak. Every year, more than $700 million is spent dealing with Salmonella outbreaks alone. Now that you know how an outbreak begins, you may be wondering how scientists stop them.
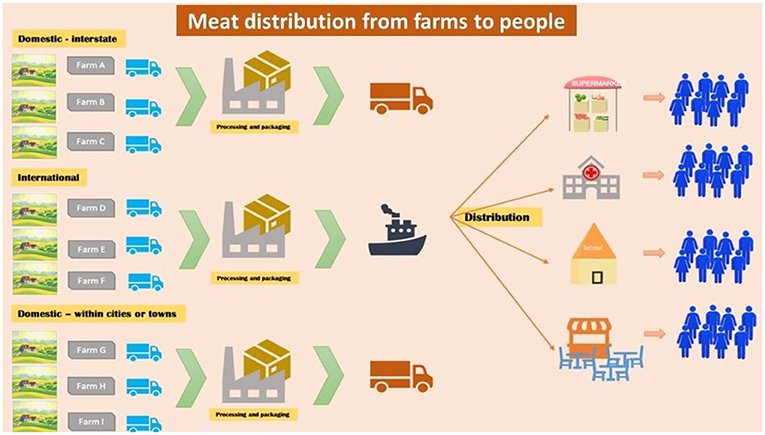
- Figure 2 - The food supply system in America is complex.
- As an example, meat could be supplied from multiple sources, some close, such as within the same city or town, and some far away, even in other countries. The meat first goes to processing and packaging centers and from there is distributed to grocery stores, schools, restaurants, and hospitals. If bacterial contamination occurs at the source or in packaging centers, this could expose large numbers of people to infection.
Since many antibiotics are not effective at treating superbugs, such as Salmonella, one strategy to limit infections is to find the source of contamination and remove the contaminated food from the supply system. This is called outbreak tracing. Outbreak tracing is very important because after finding the source, it becomes easier to contain the outbreak and stop it from spreading. A common method of outbreak tracing is taking a sample of the bacteria from an infected patient and then growing it in the lab. After this, the scientists take samples from all suspected sources, grow them also, and then compare their chemical properties to those of the patient sample. This method of outbreak tracing is both expensive and time consuming. Another problem with this method is that it is not specific enough to differentiate the type of Salmonella that caused the outbreak from other types that may simply be present in the environment.
This older method of outbreak tracing is also not as useful if the source of the outbreak is very far away. For example, this method would probably be unable to find the source of a multi-state Salmonella outbreak in the USA caused by fish from India, because it is hard to get a fish sample from India in the USA. Thanks to advances in technology, a new method called whole genome sequencing (WGS) has been developed, which can identify the sources of outbreaks even when they originate in far-away places [3].
What Is WGS, and How Is It Used for Outbreak Tracing?
Whole genome sequencing was first developed in the mid-1990s [4]. The basic discoveries that led to the development of WGS are credited to a team of scientists from the University of Cambridge. WGS does not directly find the source of the outbreak or the bacteria causing the outbreak. Instead, WGS provides data that can be used to find all this information and more. WGS works by reading the entire DNA sequence of a bacteria, which is made up of four smaller pieces called bases, represented by the letters A, T, G, and C. The entire DNA sequence of an organism is called its genome, and every type of organism has a unique genome. While the older outbreak tracing method provides a maximum of 1,000 data points to compare two bacteria, WGS offers more than 300,000 data points for comparison.
The data generated by WGS is used compare sequence differences between bacterial samples, using computer programs. To make these comparisons, scientists compare the DNA sequence of the bacteria causing the outbreak to the DNA sequences of bacteria that are possible suspects. For Salmonella, scientists would extract the Salmonella from the contaminated food that was causing the outbreak, then select the sequences, stored in the computer, of the different types of Salmonella that they think might be causing the outbreak. The computer program checks how similar each “suspect” DNA sequence is to the sample from the contaminated food source, and then generates an output that that shows how closely related the actual bacteria is to the possible suspects. The more closely the DNA sequences match, the more likely the suspect is the cause of the outbreak (Figure 3). You may find it interesting that WGS is not only used to trace bacterial outbreaks, but this method has also been used to trace people’s ancestry through their DNA.
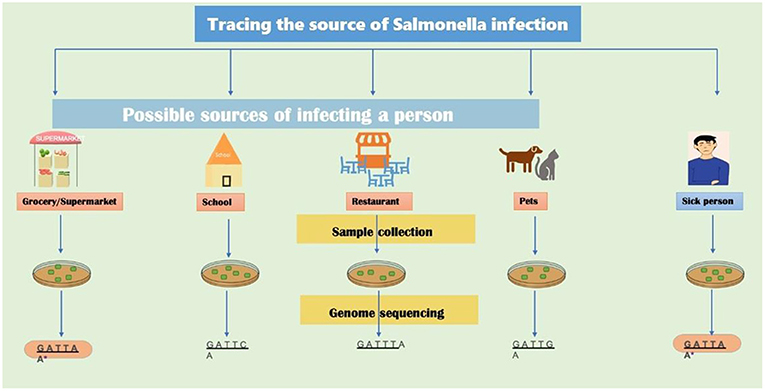
- Figure 3 - WGS-based outbreak tracing.
- To identify the source of a foodborne outbreak, samples are taken from the sick person and all suspected sources of infection. In this case, the sick person went to the grocery store, to school, and to a restaurant, and also had a pet. Salmonella is isolated from samples taken from each of these possible sources and the DNA sequences of all samples are determined using WGS. A computer program is used compare the DNA sequences. In this example, the DNA sequence from the grocery store sample and the sick person are identical, showing that the grocery store is the most likely source of infection.
Conclusion
Normally, when people get infected by a bacterium, they are treated with antibiotics. With superbugs however, antibiotics are ineffective, so these infections are extremely difficult to treat. Worse yet, if a bacterium is not treated quickly, there is a better chance for it to spread to other people. Foodborne infections with superbugs sicken 47 million and kill thousands of people annually.
Whole genome sequencing is extremely important because it can track superbugs accurately. Due to the complexity of the world’s food supply system, it has become difficult to find the source of outbreaks of superbugs, such as Salmonella. Better tracking methods, such as WGS are necessary to help scientists contain outbreaks. Better tracking can also lower the cost of tracking outbreaks and the amounts of supplies used. Good tracking also helps scientists to make better predictions about where and when an outbreak could happen, which will ultimately help us to reduce the number of superbug outbreaks that occur and save thousands of lives.
Glossary
Superbug: ↑ A bacteria that has devolved resistance to two or more antibiotics.
Outbreak Tracing: ↑ The process of tracing an outbreak to its source to stop more people from being infected.
DNA Sequence: ↑ The genetic instructions that are encoded in bacteria and other living organisms.
Bases: ↑ The four building blocks of DNA, represented by the letters A, T, G, and C.
Genome: ↑ Is the entire DNA sequence of an organism.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Alex Kidangathazhe for helping with the preparation of the figures, writing the manuscript, and the revisions. He is a seventh-grade student at the Mickelson Middle School, Brookings, SD. His hobbies include playing the piano, cooking, baking, and reading! He hopes to be a doctor 1 day and research new cures and vaccines. Writing is another one of his passions. He feels that writing gives him a better understating of the topic and helps someone else as well. He also loves traveling and getting to know new people.
References
[1] ↑ Aslam, B., Wang, W., Arshad, M. I., Khurshid, M., Muzammil, S., Rasool, M. H., et al. 2018. Antibiotic resistance: a rundown of a global crisis. Infect. Drug Resist. 11:1645–58. doi: 10.2147/IDR.S173867
[2] ↑ Brenner, F. W., Villar, R. G., Angulo, F. J., Tauxe, R., and Swaminathan, B. 2000. Salmonella nomenclature. J. Clin. Microbiol. 38:2465–7. doi: 10.1128/JCM.38.7.2465-2467.2000
[3] ↑ Antony, L., Behr, M., Sockett, D., Miskimins, D., Aulik, N., Christopher-Hennings, J., et al. 2018. Genome divergence and increased virulence of outbreak associated Salmonella enterica subspecies enterica serovar Heidelberg. Gut Pathog. 10:53. doi: 10.1186/s13099-018-0279-0
[4] ↑ Heather, J. M., and Chain, B. 2016. The sequence of sequencers: the history of sequencing DNA. Genomics 107:1–8. doi: 10.1016/j.ygeno.2015.11.003
