Abstract
DNA is a central component in the cells of all living organisms, which contains information that is critical for building the body and maintaining its biological processes. Today, owing to new technological developments, scientists can obtain DNA directly from remains of humans and other organisms that died a long time ago. The ability to read ancient DNA allows us to learn new facts about the lives of the ancients, to study genetic changes that shaped present-day organisms, and to get new insights on fascinating evolutionary questions. For example, how did the Neanderthals evolve, and why did mammoths go extinct. Owing to ancient DNA, a mysterious group of archaic humans was discovered, although almost nothing is known about their appearance. Additional important information can be revealed by looking at epigenetics, which are mechanisms that affect the way genes work. For example, through epigenetics we identified fascinating changes in the structure of the voice box (formally called larynx) of modern humans, which possibly allowed us to produce rich sounds and develop a complex language. The ancient DNA revolution provides us with new tools to learn about event that happened many years ago.
Learning to Read Ancient DNA
DNA is one of the most marvelous things in nature. Even though it is a tiny molecule, which could not be seen by the naked eye, it contains all the necessary information needed to create a living creature from a single cell and to maintain its biological processes. We can parallel DNA to a huge library whose books, which are called genes, contain the instructions for operating almost every process that takes place in our body. DNA is written in a language called the genetic code, which contains only four letters. If we know how to decipher the language of DNA, we could better understand how our body works. For this reason, several methods were developed for reading the sequence of letters of a DNA molecule, a process called “DNA sequencing.” With time, DNA sequencing became efficient, fast and relatively cheap, and today we have read the DNA sequences of tens of thousands of humans, as well as tens of thousands of other organisms.
The ability to sequence DNA opens new research possibilities that were previously regarded as science fiction. One of these fascinating possibilities is the study of ancient DNA. This means to sequence DNA that was taken from bones and other remains of organisms that died many years ago. This relies on the fact that DNA molecules are very stable and the genetic information encoded in them can be preserved for even up to a million years. It should be understood that studying the ancient past is a very challenging task. Archeologists try to investigate tools that ancient humans left behind and geologists try to understand how continents and oceans changed. But despite their many successes, it is still difficult to answer basic questions, such as: “why did mammoths go extinct?”, or “when did humans develop language?” The ability to sequence DNA of ancient organisms provides new opportunities to give answers to questions upon which we could not previously answer.
The beginning was tough, as there were many errors in the first ancient DNA sequences. But, step by step, results have become much more reliable. The first significant achievement was published in 1984, when Russel Higuchi and his collaborators reported that they managed to sequence a small piece of the DNA of a quagga, an ancient relative of the zebra, which went extinct 150 years ago. Since then, more and more ancient DNA sequences were published, and today we have access to the genetic material of hundreds of people, of archaic humans, such as the Neanderthals, of agricultural crops from thousands of years ago, of animals from the last ice age (that occurred tens of thousands of years ago) like mammoths and giant sloths, and of animals who went extinct only very recently, such as the dodo in Mauritius, the Tasmanian wolf in Australia and the passenger pigeon in North America.
The Story of the Neanderthals
Nowadays, all humans on Earth belong to the same group, to which we usually call “modern humans.” But until about 30,000 years ago, other human groups existed in parallel to us. The most well-known are the Neanderthals. Neanderthals lived for tens of thousands of years in Europe, Asia and the Middle East. At the same time, modern humans evolved in Africa. The two groups met only when modern humans started to exit Africa and spread across other continents. The nature of the encounters between the two human groups is still a mystery, but the final outcome is well-known—we are still around, while Neanderthals went extinct long ago. The Neanderthals were overall very similar to us, but still exhibited some differences. For example, they had a lower and slightly elongated skull, a slightly bigger brain, a protruding face, bigger teeth, and wider bones, indicating that they were more robust than us (Figure 1). All of this can be seen in the many Neanderthal skeletons that were found along the years. In fact, many of what we know about them is related to their skeleton, as skeletal parts survive after death much better than other tissues. However, these many skeletons could still not answer many fascinating questions—did Neanderthals develop language similar to modern humans? Did they have a culture? If so, how did their culture look like? Was the encounter between us and them violent or peaceful? Did they go extinct because of us? Did Neanderthals and modern humans produce offspring together?
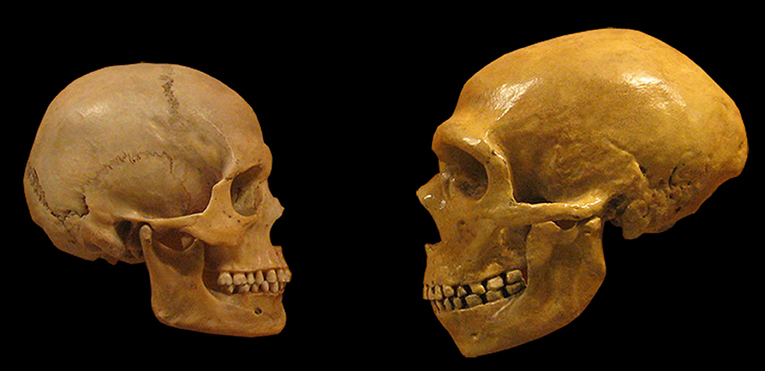
- Figure 1 - Comparison between Neanderthal (right) and modern human (left) skulls.
- Notice the thick eyebrow ridges and big jaws of Neanderthals, and the higher forehead of modern humans. The volume of the brain of both human groups was similar, but the Neanderthal brain was more elongated and flat (source: Wikimedia).
The ancient DNA revolution did not skip the Neanderthals. Over the years more and more DNA could be sequenced from Neanderthal remains found in different parts of the world. In 2014 Svante Pääbo and his colleagues published the first high-quality DNA sequence of a Neanderthal who lived tens of thousands of years ago in Siberia [1]. They found that 99.7% of the Neanderthal DNA is identical to ours. Many researchers try to understand what is the biological meaning of the 0.3% difference. For example, it was shown that Neanderthals had a slightly different version of a gene called MC1R. This gene is responsible for the color of the skin and hair. Researchers think that the Neanderthal version of this gene probably led to light skin and reddish hair. Therefore, in drawings and restorations of Neanderthals from recent years, artists usually used red as the color of their hair. Note that this information could not have been deduced from archeological findings, as we do not have Neanderthal hair and skin.
And what about the interaction between us and them? It turns out that roughly 1–2% of the DNA of modern humans from Europe, Asia, and America comes from Neanderthals. This means that Neanderthals and modern humans mated and gave birth to hybrid children. Most of us are actually offspring of these ancient hybrids.
The Denisovans—Our Mysterious Relatives
One of the amazing stories of ancient DNA takes us far away into the Denisova cave in Siberia. Researchers found there a small part of a pinkie bone and a few teeth, which they assumed belong to Neanderthals. But when they sequenced the DNA [2] it turned out to be significantly different from both Neanderthals and modern humans. Consequently, the researchers concluded that this was a new group of humans, different from anything that was previously known, and called these people “Denisovans.” This was the first time in the history of science that the existence of a new group of humans was deduced solely based on DNA. Even today not much is known about the Denisovans—neither how they looked like, nor how they lived or what were their cultural and cognitive abilities.
The DNA of the Denisovan enables us to learn a few things about these mysterious humans. Traces of their DNA were found in the genomes of native Australians (Aborigines), and native New Guineans and nearby islands. It is therefore likely that the Denisovans lived not only in Siberia but also in other places in Asia, where they met and interacted with modern humans. Denisovan DNA was found in present-day Tibetans, conferring them with the ability to live in high altitudes. Denisovan DNA was also found in present-day Inuit people (Eskimos), conferring them with the ability to live in fierce coldness. Based on these findings, it is believed that Denisovans were adapted to living in high altitudes and cold temperatures. In several regions of East Asia remains of people that are not modern humans were found along the years. It is possible that these remains are of the Denisovan, but until scientists will be able to extract DNA from them and compare it to the DNA that was found in the Denisova cave, we will not know with certainty how Denisovans looked like.
Why Did Mammoths Go Extinct?
The ancient DNA revolution provided us with interesting findings not only on humans but also on other species. A famous example is the wooly mammoth, a relative of modern elephants (Figure 2). The wooly mammoths lived in the northern parts of America and Asia. Most of them went extinct about 10,000 years ago, maybe because the climate became warmer, or because they were hunted by humans, or maybe because of both factors combined. A small community of mammoths, however, survived in a small island in the heart of the Arctic Ocean, where weather was cooler and there were no humans to hunt them. This community survived for thousands of years and went extinct <4,000 years ago. It is puzzling to think that when the pyramids were built in Egypt there were still mammoths walking on the face of the Earth.

- Figure 2 - The wooly mammoth was a relative of the Asian elephant.
- Thanks to properties, such as a tangled fur, thick layer of fat and small ears, it was acclimatized to living in cold weather (source: Shutterstock).
Thanks to the cold weather in which mammoths lived (and died), their DNA preserved well, and researchers were able to sequence DNA from mammoths from different periods. This DNA sheds new light on their extinction [3]. For example, the DNA of a mammoth from one of the arctic islands shows many harmful mutations (i.e., detrimental changes in the DNA sequence) that might have given rise to a number of diseases. This is a phenomenon that occurs sometimes in isolated islands. Since the island is small, it cannot support many mammoths, and as a result these mammoths mated with close relatives, which is known to increase the frequency of harmful mutations. With time, more and more such harmful mutations accumulated in their DNA, which probably made their population more and more prone to collapse, until their final extinction. For further reading about bringing Neanderthals and mammoths back to life see Box 1.
Box 1 - Resurrecting Neanderthals and mammoths?
Until a few years ago, the possibility of bringing ancient humans and extinct animals back to life sounded like a complete science fiction. But the ancient DNA revolution changes this. Babies and fetuses develop from the merger of two cells: an egg and a sperm, one originates from the mother and the other from the father. When these cells merge, they become a single cell called a zygote. The zygote contains DNA that came from the father (through the sperm), and DNA that came from the mother (through the egg). The information contained in its DNA determines how this fetus will develop. Theoretically, if we take a zygote from a mother elephant, extract its original (elephant) DNA, replace it with a mammoth DNA, and then re-introduce it into the mother elephant, this mother could give birth to a little mammoth (or a hybrid of an elephant and a mammoth). Today, there are a number of research groups that investigate this option. The famous American geneticist George Church already declared: “I can create a Neanderthal baby, I just need a willing woman.” But, as of today, the researchers still face several technical obstacles along the way. An elephant’s womb is slightly different than that of a mammoth, and this structural difference might affect the fetus. Also, it is still technically challenging to modify an elephant DNA and turn it into a mammoth DNA. Finally, development might be affected by epigenetic signals and therefore such a feat must take into account also epigenetic information. Finally, we should ask ourselves whether resurrecting extinct animals, and specifically extinct human groups, is the right thing to do, morally and ethically. These are questions that should be addressed today, as in the foreseen future all technological barriers are expected to be overcome, and bringing Neanderthals and mammoths back to life would be technically feasible.
Epigenetics—A New Way of Understanding the Past
When a tadpole matures into an adult frog, its DNA does not change. Yet, a frog looks very different from a tadpole. A tadpole has a tail, has no legs and it breaths underwater. In contrast, a mature frog has no tail, has four legs and it breaths on land. How could the same DNA create animals that are so different? The answer lies in what is called gene regulation. Let us go back to paralleling DNA to a huge library, and genes to books with instructions. Now, imagine that it is possible to use a red rope to block access to several parts in this library. As a result, although some books are present in the library, no one can access them and read them. In tadpoles, the “frog genes” are blocked, whereas in frogs the “tadpole genes” are blocked. An important component of gene regulation is chemical changes in the DNA or in the way that it is packed in the cell. These changes are called by the general name “epigenetics” (Figure 3).

- Figure 3 - The effects of genetic and epigenetic changes.
- 1: Changes in the sequence of letters comprising the genetic code result in a change of the color of the corn of the unicorn from blue to red. This is a genetic change. 2: There is no change in the sequence of the DNA but there is a change in gene regulation. The red circles denote DNA that underwent a modification called “DNA methylation” (M), which makes the gene “inaccessible.” As a result, the corn’s color changes to green.
Since the DNA of modern humans is very similar to that of archaic humans, researches think that most of the differences between the groups do not result from changes in the DNA itself, but from changes in gene regulation. The problem is that it is very difficult to reconstruct patterns of gene activity only by looking at the DNA sequence. In 2014, our research group, in collaboration with Prof. Eran Meshorer, achieved a breakthrough [4] and developed a method to reconstruct epigenetic patterns in ancient DNA and, consequently, to reveal how genes were regulated when the ancient organisms lived. More specifically, we found how to reconstruct epigenetic patterns which are called “DNA methylation,” that enable us to understand which genes are active and which are silenced. We reconstructed the methylation patterns of the Neanderthal and the Denisovan, and found genes whose genetic code is identical among different human groups but their epigenetic pattern is very different, and therefore the genes function in a different way.
In this way we obtained a list of genes that act differently in modern and in archaic humans. This is a fascinating list, including many genes connected to brain function and neurological diseases. Many other genes in this list are connected to the structure of our voice box. When comparing human to chimpanzee, it can be observed that the voice box of the chimpanzee is located higher along the throat compared to our voice box, and that their face protrudes forward compared to ours. We know from non-human great apes that were raised in captivity that even if we teach them to speak, they will not vocalize the words but rather express themselves by using the sign language and other gestures. It is believed that the reason for this is that they are incapable of producing the same range of sounds that we can produce, because of the different structure of their voice box. Remarkably, the list of genes whose activity pattern is unique to modern humans, we identified genes that are responsible for these anatomical differences—their different activity lead to flattening of our face and lowering of our larynx. Therefore, Neanderthals and the Denisovans might have had a higher voice box, possibly even leading to differences in sound production.
The reconstruction of epigenetic patterns opens up new possibilities in the study of the past. For example, it is known that, in contrast to the DNA itself, epigenetics might change in response to changing environment. Therefore, in principle, it should be possible to use epigenetic patterns in order to reconstruct the environment in which ancient humans lived. For example, did they suffer from hunger? Emotional stress? Cold? Preliminary clues suggest that Neanderthals and Denisovans lived in a similar dietary condition as that of hunter-gatherers. The ability to sequence ancient DNA, and to reconstruct gene regulation, is a real revolution. Using them, we can study new details on lost chapters in the history of the human race and to understand how we became who we are today.
Glossary
DNA: ↑ A long molecule that is present in every cell in our body. This is the hereditary material, and it contains important information needed to build the body and maintain its life processes. The DNA is a long chain built from four types of building blocks.
Molecule: ↑ The smallest division of matter, built from a number of atoms. Most materials around us are, eventually, made from many molecules. Examples of well-known molecules are: water, carbon dioxide, salt, and DNA.
DNA Sequencing: ↑ Reading the sequence of the building blocks (“the letters”) that comprise a DNA molecule. This procedure is usually performed in order to decode the information that is stored in the DNA sequence.
Ancient DNA: ↑ DNA that was retrieved from the remains of humans or other organisms that lived in the past.
Neanderthal: ↑ A group of archaic humans that was widespread in Europe, Asia and the Middle East, and went extinct about 30,000 years ago. They are named after the Neander valley in Germany, where their first remains were found.
Denisovan: ↑ Another group of archaic humans that was widespread in Asia and went extinct. No complete skeletons of the Denisovans were found as of today. They are named after the Denisova cave in Siberia (Russia), where their first remains were found.
Epigenetics: ↑ Regulatory processes that block or facilitate access to genes along the DNA. A change in these regulatory processes could lead to changes in appearance or behavior, even if the DNA molecule itself does not change at all.
Hunters-Gatherers: ↑ Human populations that obtain food by hunting animals and gathering food edible parts of plants and fungi. Most often, hunters-gatherers are vagabonds. In the past, all humans were hunters-gatherers, but after the Neolithic revolution they settled in permanent settlements and became farmers who grow their own food.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
[1] ↑ Prüfer, K., Racimo, F., Patterson, N., Jay, F., Sankararaman, S., Sawyer, S., et al. 2014. The complete genome sequence of a Neanderthal from the Altai mountains. Nature 505:43–9. doi: 10.1038/nature12886
[2] ↑ Meyer, M., Kircher, M., Gansauge, M. T., Li, H., Racimo, F., Mallick, S., et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338:222–6. doi: 10.1126/science.1224344
[3] ↑ Rogers, R. L., and Slatkin, M. 2017. Excess of genomic defects in a woolly mammoth on Wrangel island. PLoS Genet. 13:e1006601. doi: 10.1371/journal.pgen
[4] ↑ Gokhman, D., Lavi, E., Prüfer, K., Fraga, M. F., Riancho, J. A., Kelso, J., et al. 2014. Reconstructing the DNA methylation maps of the Neandertal and the Denisovan. Science 344:523–7. doi: 10.1126/science.1250368
