Abstract
Bacteria are single-celled organisms that live out their lives at a microscopic scale. We can find bacteria everywhere we look for them, including inside of our own bodies. Bacteria are incredibly diverse and come in many shapes and sizes. They also vary widely in how they live and grow. Some bacteria grow very quickly and others grow slowly. We wanted to measure the growth of many different types of bacteria in the environment. Unfortunately, some species of bacteria are very difficult to grow in the laboratory. To get around this, we designed a method to predict how fast a type of bacteria can grow, just from its DNA. This way, if we have the DNA of a bacterial species, we can measure its growth even if we cannot get it to grow in our laboratory.
Some Bacteria Grow Quickly, Others Not So Much
Most life on our planet is tiny. Bacteria are single-celled organisms that live out their lives at a microscopic scale. We can find bacteria everywhere we look for them, including inside of our own bodies. Even more impressive than their abundance, bacteria are incredibly diverse and come in many shapes and sizes. They also vary widely in how they live and grow. For example, some species of bacteria can reproduce very quickly, but others take a long time to reproduce. Some may even take thousands of years [1]! As scientists, we want to know why some bacteria grow quickly and others slowly. For example, if a species of bacteria can make you sick and it also grows quickly, that could be a dangerous combination!
Bacteria reproduce via a process called binary fission. This means that a single cell grows, reproduces its DNA, and eventually divides in two to create two “daughter” cells (Figure 1). Basically, bacteria clone themselves, creating an almost identical copy. The rate at which bacteria divide varies widely. Some fast-growing species can divide in under 10 min. On the other hand, some slow-growing bacteria may wait days or more between divisions. For example, Salmonella enterica, a nasty bacterium that can give you food poisoning when it grows in your gut, can divide every 20 min. On the other hand, Prochlorococcus is one of the most abundant organisms growing in the world’s oceans [2] and takes a day or more to complete a single division. We call the time that it takes a bacterial cell to divide the doubling time (Figure 1). We can measure the rate at which a population of bacteria grows in terms of its doubling time. When bacteria grow quickly, that means a short doubling time. When bacteria grow slowly, that means a long doubling time.
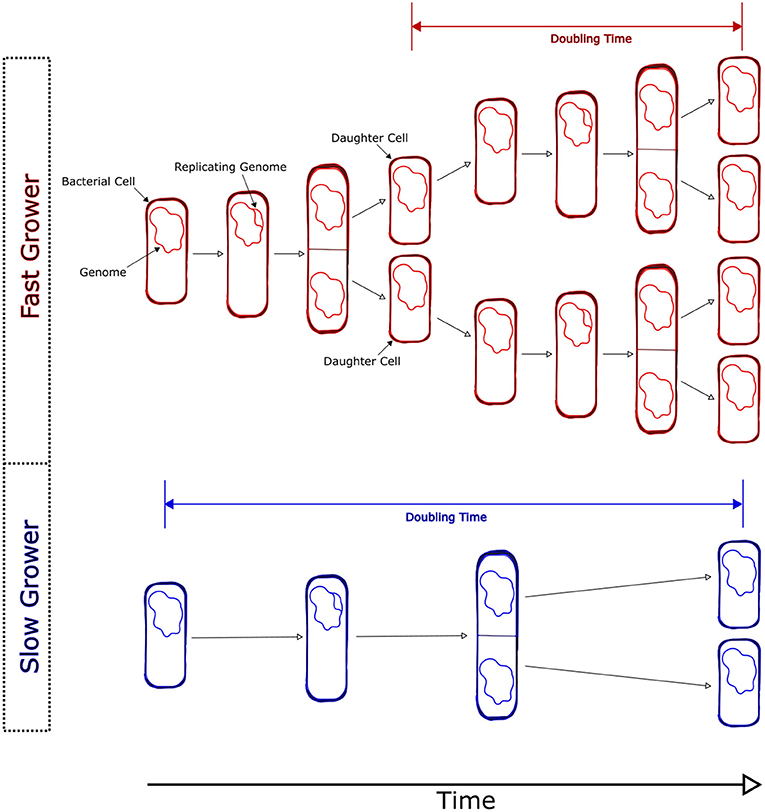
- Figure 1 - Bacteria reproduce via binary fission.
- As cells grow, they increase in size and eventually begin replicating their genomes (DNA). Eventually, a bacterial cell splits in two, with each “daughter” cell receiving a single copy of the genome. The stretch of time between divisions is called the doubling time. Cells that grow quickly will have a short doubling time, whereas cells that grow slowly will have a long doubling time.
Bacteria will not always grow quickly—they must be given the right conditions. When food is plentiful, bacteria will grow quickly, with short doubling times. As resources become scarce, bacteria may begin to divide more slowly, resulting in longer doubling times. This is why you have to worry about your food going bad—for tiny bacteria, any human food is a LOT of food and so they begin to divide rapidly. On the other hand, we do not often worry about large numbers of bacteria accumulating on most surfaces in our homes because usually there is not much available there for them to eat. Licking the wall is gross, but it probably will not make you sick like eating spoiled food would!(But do not go licking walls after reading this! You never know what might be hanging around...).
The amount of food available is not the only thing limiting a bacterium’s growth. Some bacteria simply grow faster than others. That is, different species of bacteria have different minimum doubling times. Minimum doubling time is the time it takes for the number of cells to double under optimal conditions. That is, when the bacteria have plenty of food, how fast could they possibly grow? Think of it this way: if you are hungry and thirsty, you might not be able to run as fast as if you were well-fed and hydrated. But no matter how much you eat and drink, you will never run as fast as a cheetah. Some species have evolved to grow (or run) faster than others.
How Do You Measure Growth Without Growing Microbes?
Unfortunately, we cannot measure the growth rates of many bacteria because we do not know how to grow most of them in the laboratory. It is not easy to figure out what a bacterial species’ favorite food is, or what temperature it likes to grow at, or any of a species’ various other preferences (salinity, pH, etc.). Figuring these things out can be expensive and time consuming, so we only know the growth conditions for a relatively small number of bacteria (around 14,000) compared to the total number of bacterial species estimated to exist (around 400,000) [3]! The bacteria that we cannot grow in the laboratory are a big mystery, and figuring out how fast they grow is the first step in figuring out what they could be doing in the wild.
What can a scientist do? Sometimes we cannot grow a species of bacteria in the laboratory, but we can still learn a lot about it. How, you ask? We can collect these bacteria from their natural environments and examine their DNA, without ever having to grow them. We will not get into the details here, but the main point is that even when we cannot grow bacteria in the laboratory, we can sometimes still recover its DNA sequence. The DNA sequence of bacteria can tell us how fast that bacterial species can grow.
Clues to Growth in Bacterial Genomes
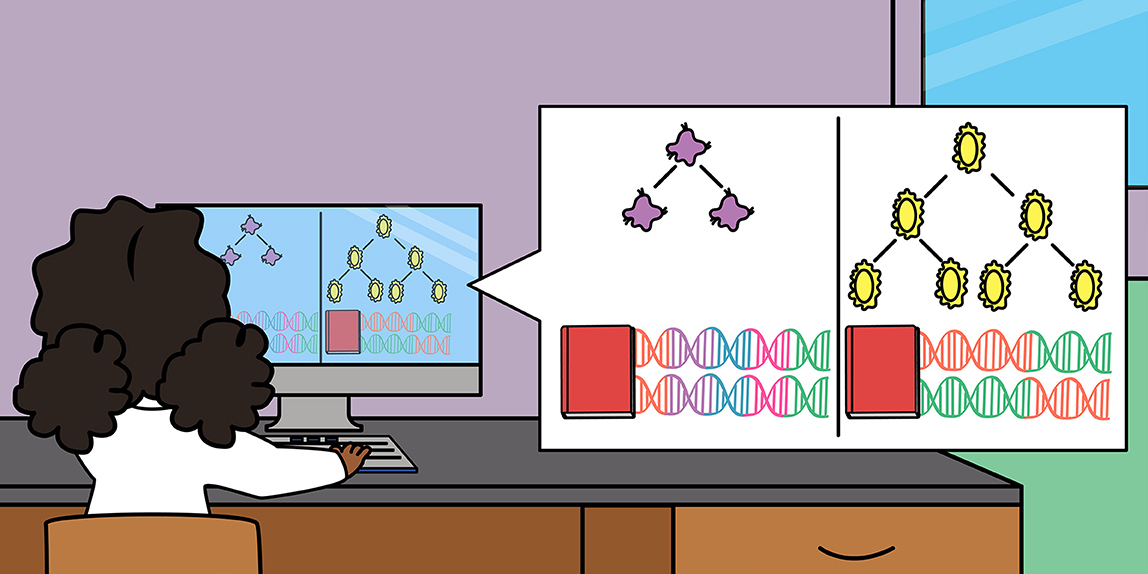
Ok, so how can we tell how fast a bacterium can divide just from its DNA sequence? Well, before we can tell you how to predict growth rate from DNA, we need to tell you a little bit about how DNA works. Every living thing is made up of cells, and each of these cells has DNA, which provides the “instruction manual” for making an organism—the instructions needed for the cell to live and grow. We call the collection of DNA inside a cell the cell’s genome. Genes are stretches of DNA that each tell a cell how to make a specific protein. Proteins are molecules that make up the machinery of a cell.
Ok, that is a lot to take in! Here is an easy way to think about it. The genome is like an instruction manual for how to build the cell, and each gene is like a single “chapter” in the manual, containing the instructions to make one piece of the cell (Figure 2A). So, if genes are like the chapters of the instruction manual, what are the words? The DNA alphabet contains four letters: A, T, G, and C. Each letter represents a different type of molecule that can get incorporated into the longer DNA chain. All genetic “words” are three letters long and we call them codons. Each word codes for an amino acid, one building block of a protein, and together a long sequence of words (the gene, or “chapter”) is used to build a specific protein with a specific function in the cell.
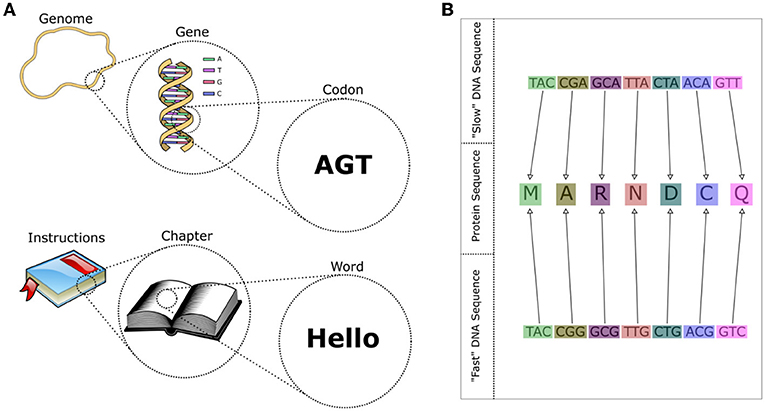
- Figure 2 - (A) A genome can be thought of as an “instruction manual” to build a cell, with each gene being a “chapter,” and each codon a “word.” All codons are made up of three letters—all words in “DNA language” are three letters long.
- (B) The language of DNA is redundant, which means that multiple words (codons) have the same meaning. The same protein sequence can be produced by different DNA sequences. Some of these sequences are easier for the cell to produce quickly than others, allowing for faster growth.
Just like English or any other language, the language of DNA is redundant—different words can mean the same thing. Different codons can code for the same amino acid (protein piece) (Figure 2B). But, just like you, the cell prefers to use certain words more than others. For example, most people would rather say “I feel hungry” than “I am famished,” even though the two sentences have the same number of letters. For most of us, the word “hungry” comes to mind faster than the word “famished.” Even if a word is in our vocabulary, we might not like to use it, especially if we are trying to communicate quickly. All species of bacteria have the same codons in their “vocabulary,” but each species prefers to use some codons more than others. When a cell wants to grow quickly, it is easier for it to use its preferred set of codons to build proteins. Over time, this preference means that natural selection favors the use of a smaller vocabulary in faster-growing species. We can measure the size of a cell’s preferred vocabulary to predict how fast it can grow—a smaller preferred vocabulary means a faster grower (and a shorter doubling time).
We built a computer model [4] that measures the size of a bacterial species’ codon “vocabulary” to predict the growth rate of those bacteria (Figure 3). This allows us to estimate how fast the bacteria can grow, even if we cannot grow them in the laboratory, as long as we have sequenced the species’ genome. This means we can measure the growth rate of many more types of bacteria than we could before.
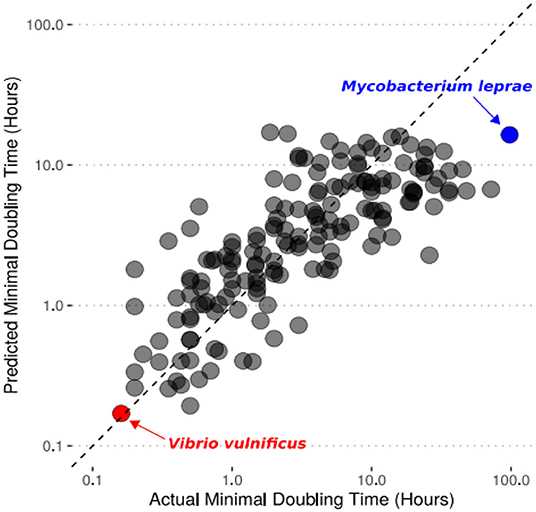
- Figure 3 - Our computer model’s predictions for the doubling time of a species are similar to the actual minimal doubling times measured with lab experiments.
- The dashed line is the x = y line, which represents a perfect prediction. Each point represents a different bacterium that we tried to predict the growth rate of. We highlighted Vibrio vulnificus in red because it is the fastest-growing bacteria we predicted. We highlighted Mycobacterium leprae in blue because it is the slowest-growing bacterium we predicted.
Growing Microbes Is Difficult When They Grow Slowly
We found using our computer model that the bacteria we grow in the laboratory mostly grow really quickly. Think about it, would you want to wait around for a bacterium to grow? Even worse, many of these slow-growing bacteria are also really picky, so it is hard to figure out the conditions they require for growth. Altogether, it is very time consuming and tedious to grow slow-growers in the laboratory. This means that we actually do not know very much about slow-growing bacteria, at least compared to their fast-growing cousins.
When we predicted the doubling times of bacteria taken directly from the environment, we saw that their doubling times were much longer than those that we already know how to grow in the laboratory. This showed us why scientists have mostly studied fast-growing bacteria in the past. Scientists, for the most part, have limited time and resources. They will sometimes work with the easiest set of bacteria rather than bacteria that are most common in the environment. Some scientists, though, are working very hard to learn more about slow-growing bacteria. It will be challenging to grow these bacteria in the future, but much remains to be learned about their biology. This is especially true since some environments, like the oceans, are mostly full of slow-growers.
Thinking Ahead
DNA can tell us a lot about how bacteria live. In this article we talked about how DNA can tell us about growth rates, but there are many other interesting things to study about bacteria. What else can DNA tell us about bacteria? What other traits of bacteria might we want to study? If you were going to use DNA to predict something about bacteria, what would it be?
Funding
JW was supported by a postdoctoral fellowship in marine microbial ecology from Simons Foundation Award 653212. We also acknowledge support from Simons Foundation Collaboration on Computational Biogeochemical Modeling of Marine Ecosystems (CBIOMES) Grant 549943 (to JF) and US NSF Division of Ocean Sciences (OCE) Grant 1737409 (to JF).
Glossary
Binary Fission: ↑ The reproductive process for bacteria, during which a cell splits into two identical “daughter” cells.
Doubling Time: ↑ The amount of time between cell divisions. A faster-growing bacterium has a shorter doubling time and a slower-growing bacterium has a longer doubling time.
Genome: ↑ All of a cell’s DNA. The “instruction manual” for the cell.
Gene: ↑ A stretch of DNA that provides the instructions to build a protein.
Proteins: ↑ Important pieces of the cell that together do much of the work of life.
Codon: ↑ A single DNA “word” that codes for a specific piece of a protein.
Amino Acid: ↑ A building block of a protein. Each is coded for by one or more codons.
Computer Model: ↑ A program we build on the computer to better understand and predict how biology works.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Original Source Article
↑Weissman, J. L., Hou, S., and Fuhrman, J. A. 2021. Estimating maximal microbial growth rates from cultures, metagenomes, and single cells via codon usage patterns. Proc. Natl. Acad. Sci. U.S.A. 118:e2016810118. doi: 10.1073/pnas.2016810118
References
[1] ↑ Jørgensen, B. B., and D’Hondt, S. 2006. Ecology-A starving majority deep beneath the seafloor. Science. 314:932–4. doi: 10.1126/science.1133796
[2] ↑ Partensky, F., Hess, W. R., and Vaulot, D. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106–27. doi: 10.1128/MMBR.63.1.106-127.1999
[3] ↑ Lewis, W. H., Tahon, G., Geesink, P., Sousa, D. Z., and Ettema, T. J. 2020. Innovations to culturing the uncultured microbial majority. Nat. Rev. Microbiol. 22:1–6. doi: 10.1038/s41579-020-00458-8
[4] ↑ Weissman, J. L., Hou, S., and Fuhrman, J. A. 2021. Estimating maximal microbial growth rates from cultures, metagenomes, and single cells via codon usage patterns. Proc. Natl. Acad. Sci. U.S.A. 118:e2016810118. doi: 10.1073/pnas.2016810118