Abstract
Do you know how much information is hidden inside your own cells? It is a huge amount. Are you curious about how scientists decipher and handle this large volume of data? Computer science and mathematics have helped biologists to create a new science to analyze, organize, and understand biological data: bioinformatics. Bioinformatics allows us to manage huge amounts of biological data and make sense of it. In other words, bioinformatics allows us to explore the mysteries of life and find answers to complex questions about the functioning of living beings. In this article, we are happy to tell you what this fascinating new field is and what it is used for. We think you will like it!
Bioinformatics: Interpreting Biological Symbols
You might be surprised to learn that each cell of your body contains a “handbook” with the instructions to build and maintain your entire body—all kept in a space of 4–6 μm (a μm is one millionth of a meter)! This information is stored in the DNA, which is found in almost all living organisms and is the “universal language” of life. In human cells, this language consists of more than six billion “letters”. How is it possible for scientists to study and interpret the enormous amount of information written in the DNA? This difficult task led to the birth of a new science, called bioinformatics [1].
The Handbook of Life
Scientists call the DNA handbook, or the entire set of DNA instructions found within each cell, the genome (Figure 1). The handbook is divided into “chapters” known as chromosomes. Each species has a specific number of chapters, or, in other words, each species has a set number of chromosomes. For example, humans have 23 pairs of chromosomes and tomatoes have 12 pairs. DNA is a long molecule made up of a sequence of four “letters” corresponding to four chemical compounds: adenine (A), guanine (G), cytosine (C), and thymine (T). Here is an example of what a DNA sequence looks like: ATGGTCCCATGCTAGCTAGCTATCGATGCTACGTACGTAG
CATAAATCGCGATAGCTA.

- Figure 1 - You can think of your genome as a handbook containing all the instructions needed to create your body and allow it to function.
- Each “chapter” of this handbook is a chromosome, and humans have 23 pairs of them. Within each chapter, the sentences containing the actual instructions are called genes (red).
Combinations of those four letters are used by cells to know how to behave. Everyone’s genome is a little bit different, and the sum of these differences leads to the unique way that you look and can even influence how you think and act.
So, how can scientists read and understand the genome? “Reading” a genome means knowing the sequence of letters that make it up. For this purpose, scientists use a technique called genome sequencing (you can read more about genome sequencing in this Frontiers for Young Minds article). But knowing just the sequence is like having a book written in an unknown language that must be deciphered. First, bioinformaticians seek to identify specific sentences within the genome, referred to as genes. Each gene contains the information needed to make a specific protein. In fact, every time the cell wants to do something, it goes back to the handbook, selects the necessary lines of text (i.e., the genes) and uses them to build proteins. Proteins are the workers of the body, each with its own special job to do. Proteins can help to move things around inside cells, can help to build cells, and more. Proteins can interact with each other or with other chemical compounds to build muscles, hair, and nails, for example.
Proteins are molecules made up of various assortments of 20 chemical compounds, the amino acids. An amino acid can be thought of as a building block, such as a LEGO brick, and a protein as a particular LEGO masterpiece. Depending on how you assemble the blocks, you can get specific creations. Similarly, a cell assembles amino acids to obtain a particular protein (you can read more about proteins in this Frontiers for Young Minds article). Every time the cell needs a protein, it translates the instructions encoded in the corresponding gene into a particular sequence of amino acids. To perform this translation, the cell applies a set of rules known as the genetic code. The genetic code allows scientists to decode the DNA sequence just as the three translations on the Rosetta Stone helped people to understand Egyptian hieroglyphics (Figure 2) [2]. Your genome contains around 20,000 genes that can encode at least 80,000 proteins that, together, regulate your body’s function.
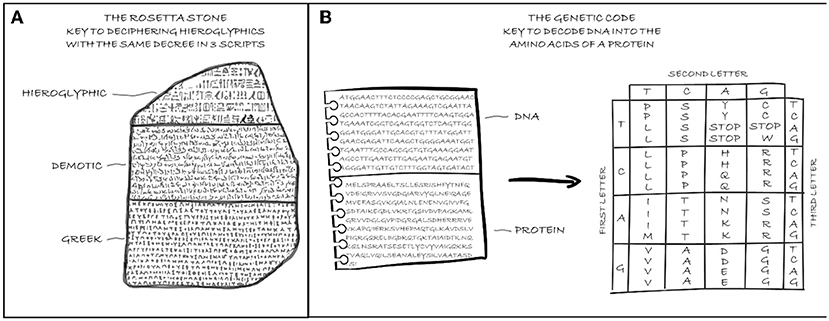
- Figure 2 - (A) The Rosetta Stone was the key to deciphering hieroglyphics.
- (B) The genetic code is the key to decoding the DNA sequence to know which protein it codes for.
In summary, both genes and proteins can be represented by a string of characters, and the exciting role of bioinformaticians is to study these sequences to decipher their secrets. If this interests you and you cannot wait to become a bioinformatician, do not worry! All you need is a laptop to get started! The first thing bioinformaticians do is to get familiar with text files on computers where DNA or protein sequences are stored. Then, with the help of some software, they can navigate, decipher, and interpret this world made of letters.
The Basic Task in Bioinformatics: Sequence Comparison
Scientists call DNA and protein sequences biosequences. Your own biosequences can be compared with all other known biosequences to infer a common evolutionary descent or a common structural function. But what does it mean to compare biosequences? It means using computer programs to line them up in pairs, to match as many letters as possible (Figure 3). The score of this alignment is given by the sum of the matches (matching symbols), the mismatches (non-matching symbols), and the gaps (additions or deletions relative to the other sequence). In other words, aligning two sequences means to identify regions of similarity [3]. Protein sequences can be compared, too. There are bioinformatics tools that allow scientists to translate a DNA sequence into a protein sequence, and then proceed with the alignment and analysis.

- Figure 3 - (A) Pairwise alignment between two protein sequence.
- The identical (matches) and different (mismatches) amino acids are highlighted in the two sequences as well as the gaps (no matches). (B) A new sequence, defined as a “query”, can be compared against a whole database of known sequences. This operation, known as database searching, returns a list of pairwise alignments and can tell scientists whether the sequence is already known or how similar it is to known sequences.
Instead of doing pairwise comparison, bioinformaticians can also use tools that allow them to search a database for a specific sequence. For example, if they have a sequence and want to know what type of protein it codes for or what its function might be, specific software can compare that new biosequence with all known biosequences stored in specific databases. The software will return pairwise alignments of all known sequences that are similar to the unknown sequence. The greater the similarity, the more likely the genes or proteins perform the same functions inside the cell.
Imagine that you have just obtained a new sequence from an experiment. You know nothing about its properties or what its role might be in a living cell. So, the first thing you can do is compare your unknown sequence (sequence X) with a large database of sequences with known properties. If sequence X is similar to sequence Y, which is known to move chemical compounds in or out of the cell, for example, then you can assume that sequence X might also be involved in the transport of specific molecules. Overall, sequence comparison is the first and most informative step in the analysis of newly determined sequences.
Bioinformatics: An Ever-Evolving Science
Since 1978, when the term “bioinformatics” was introduced, the field has seen explosive growth— especially in the last 20 years. Technology has evolved to make bioinformatics even more powerful. Data are securely stored in supercomputers and organized into databases. Data are also analyzed by increasingly efficient computer programs developed to cope with a growing volume of information, often called “big data”. The use of supercomputers and some forms of artificial intelligence can help to extract even more meaningful information from biological data. As bioinformatics continues to develop, this science will help us to better understand how living organisms work and to plan all kinds of exciting ways to help people, from defending humans against diseases to helping plants adapt to climate change.
Glossary
Bioinformatics: ↑ The science of storing large amounts of complex biological data and analyzing it to find new insights.
Genome: ↑ The set of DNA instructions found in a cell.
Genome Sequencing: ↑ The process that determines the order of the chemical building blocks, called bases, that make up the DNA molecule.
Genes: ↑ Functional units of heredity that carry the information that specifies the characteristics transmitted from generation to generation.
Proteins: ↑ Large and complex molecules that play many important roles in living organisms.
Amino Acids: ↑ Molecules used by all living things to make proteins.
Genetic Code: ↑ The set of rules used by cells to translate information encoded within DNA into proteins.
Biosequence: ↑ A series of biological molecules, such as DNA and proteins.
Acknowledgments
The authors thank all members of the GenoPom Lab for stimulating discussions during the preparation of this article. Special thanks go to our little friend (13 years old) Elizabeth Villmer for giving us feedback from a younger audience’s perspective.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
[1] ↑ Luscombe, N. M., Greenbaum, D., and Gerstein, M. 2001. What is bioinformatics? An introduction and overview. Yearbook Med. Informat. 10:83–100. doi: 10.1055/s-0038-1638103
[2] ↑ Caskey, C. T., and Leder, P. 2014. The RNA code: Nature’s Rosetta Stone. Proc. Natl. Acad. Sci. U. S. A. 111:5758–9. doi: 10.1073/pnas.1404819111
[3] ↑ Rosenberg, M. S. 2009. Sequence Alignment: Methods, Models, Concepts, and Strategies. Berkeley, CA: University of California Press.

