Abstract
Petroleum is the most-used energy source in the world. However, as you probably know, petroleum is a fossil fuel that is very harmful to the environment, in addition to not being renewable. Biofuels are a type of fuel produced from plant material. Biofuels are considered an excellent alternative energy source because they are less polluting than fossil fuels. However, biofuel production is expensive. Therefore, scientists are working on many strategies to reduce biofuel costs, particularly using computers to discover new biotechnological products, or improve existing ones, to produce more biofuel with fewer costs. In this article, we will tell you how computers can be used to improve biofuel production.
Biofuels: An Environmentally Friendly Alternative
Petroleum-based products are widely used to power our cars, to heat our homes, to generate electricity, and to make plastics. Fossil fuels, like petroleum oil, are buried deep within the Earth. They take millions of years to form and their formation depends on high-pressure environments and dead organisms like plants, algae, bacteria, and animals (including dinosaurs). When burned, fossil fuels release carbon dioxide. Therefore, the more petroleum burned, the more carbon dioxide is released into the atmosphere, contributing to global warming [1].
Biofuels may be a better source of energy. Biofuels can be produced from plants, such as corn, sugarcane, and soy. Because they come from plants that we can continue to grow, biofuels are considered renewable and sustainable, which means we can produce this kind of energy continuously [2]. However, biofuel production is expensive. Many complex processes are needed to create biofuels from plant biomass. So, many people still believe that petroleum is the more cost-friendly choice, but they are ignoring the long-term environmental issues.
For years, scientists have been working to improve biofuel production. For instance, to produce biofuel from sugarcane, sugar is extracted from cane juice and used to produce bioethanol (a type of fuel), through a process called fermentation. However, lots of waste biomass and sugar are left over after the extraction process. A recent study in Brazil estimated that, if the leftover sugar was extracted, biofuel production could be doubled [3]! Biofuel produced from sugarcane biomass is called second-generation biofuel.
Second-generation biofuel production involves many steps (Figure 1). Saccharification is a crucial step: enzymes are used to break down the leftover sugarcane biomass. Enzymes are proteins, and like all proteins they are made of chains of subunits called amino acids (that are made by atoms). Enzymes speed up chemical reactions, like breaking down other substances. The sugarcane waste and enzymes are mixed together in a large tank, where the enzymes break down the waste to release sugar. Different enzymes have different abilities to release sugar from sugarcane waste [5]. Improving the less-efficient enzymes could be a good strategy to improve this stage of biofuel production. Computers can be used to detect the most essential characteristics of efficient enzymes, then these characteristics can help scientists to design enzymes that are more efficient.
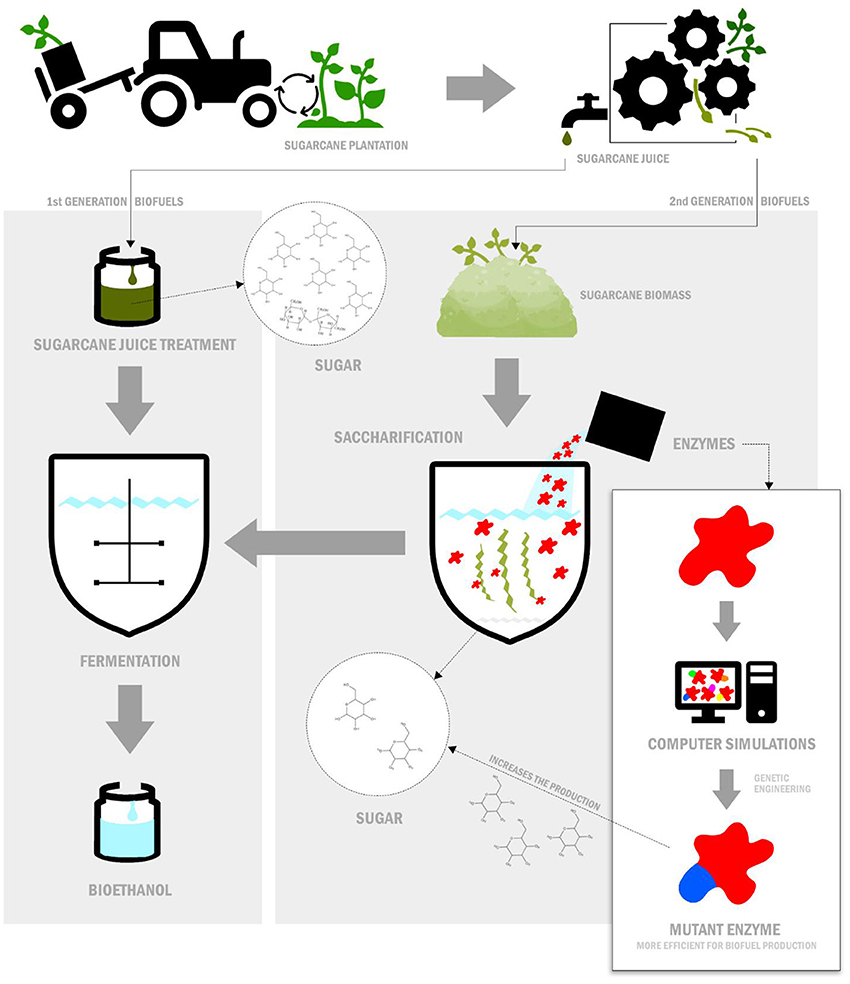
- Figure 1 - Production of first- and second-generation biofuels.
- First-generation biofuels are easily produced (for example, from the sugarcane juice). Second-generation biofuels are produced from the plant biomass left over from making first-generation biofuels. This process uses enzymes to break down the leftover plant biomass to release sugar molecules (called saccharification). Converting sugar into bioethanol is called fermentation. Computer simulations, used together with genetic engineering, can produce enzymes that are good at saccharification, so that biofuels can be produced more easily.
How Are Computers Making the Difference?
Scientists can use genetic engineering to generate enzymes that are more efficient at helping chemical reactions happen faster. Genetic engineering is complicated. It involves mutating the structure of an enzyme and studying how the mutations affect the enzyme’s function. There are zillions of possible mutations and combinations of mutations—for instance, an enzyme with 400 amino acids could be given 19400 amino acid mutations! A scientist could not possibly make and test all of these mutations! Instead, computers can be used to simulate mutations, pointing out the most promising ones to test in lab experiments. Special computer programs can simulate the structures of molecules like enzymes, based on their DNA sequences. Some software even uses graphics cards (traditionally used for running games) to show the functions of mutant enzymes as a movie [4]!
Algorithm: A Complex Word for a Simple Thing
Computers are powerful, but they also have limitations. If we want a computer to do something, a step-by-step procedure must be created. This procedure is called an algorithm. For example, to design a better enzyme, we must first understand the structure of the original enzyme. Every enzyme has a unique signature pattern, almost like a fingerprint, based on the types of atoms it contains. Enzymes are composed of several atoms connected by chemical interactions. Atoms are very small particles, and the distances between them are also tiny. The types and locations of atoms in an enzyme determine the enzyme’s shape, function, and how efficient it is at biofuel production. Thus, we created an algorithm to analyze each atom and its neighbors, to give us the enzyme’s signature pattern. Using computers, we can represent the signature patterns of enzymes mathematically, using a list of numbers [6]. Then, using simple equations, we can calculate the distance between the numbers to determine how similar enzymes are to each other. Similar enzymes will have similar signature patterns and might have similar functions.
Then, we proposed possible mutations for our non-effective enzymes to make their signature patterns more like those of the efficient enzymes. To do this, we calculated the distances among the enzymes’ signatures. This can be a little confusing. Look at Figure 2 to understand better. Remember: similar enzymes will have similar signatures. Also, enzymes with similar signatures will be closer than enzymes with different signatures. Therefore, we can use the distances to compare efficient and non-efficient enzymes. For example, imagine that the blue enzyme is a known efficient enzyme for biofuel production (do not worry, many other scientists probed that before). Additionally, pink and green enzymes are two mutants produced by genetic engineering (unfortunately, we do not have additional information about them). Based on our algorithm, we could suppose that the pink enzyme is the most efficient mutant for biofuel production because its signature point is closer to the blue enzyme’s signature point (the efficient one) than the green’s point (the other mutant).
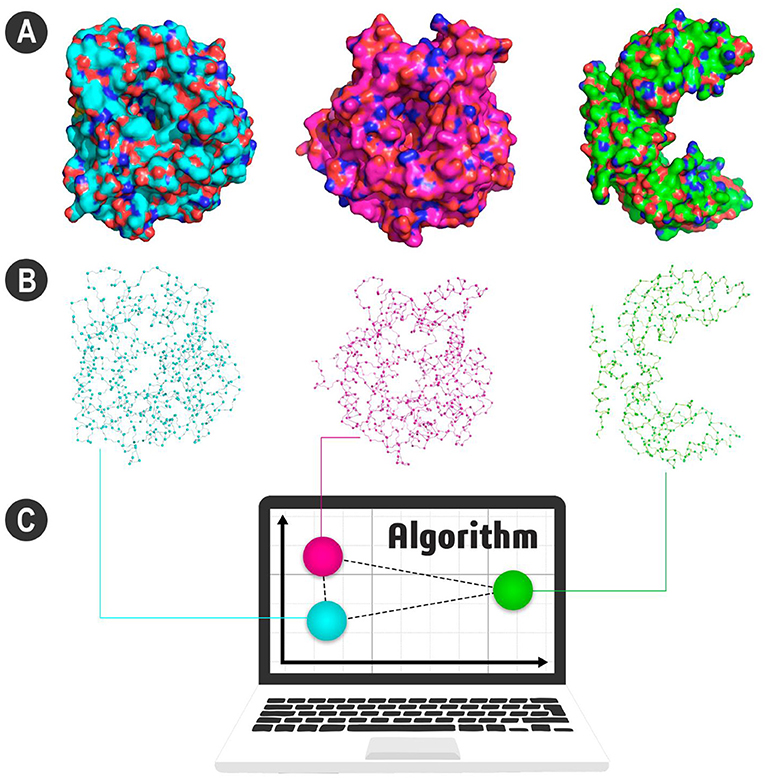
- Figure 2 - (A) Visualization of three enzymes.
- Note that the blue and pink enzymes have a similar shape. Both are enzymes used for biofuel production, but the green enzyme, with a very different shape, is not. (B) Enzymes’ representation as atoms. Some of the main atoms in each enzyme are shown connected to their neighbors by lines. Our algorithm counts the neighboring pairs and converts this into a set of numbers. All these numbers give us the signature pattern of the enzyme. (C) Here, each signature is represented as a sphere. Look how the spheres of similar proteins are closer. Therefore, the blue and pink enzymes have more similar signature patterns, different from the green enzyme. Figure generated using PyMOL (https://pymol.org).
How Do Computers Compare Mutant Enzymes?
Suppose that each enzyme’s signature is like a star in the sky. Stars can be grouped in constellations. How do we know which stars belong to the same constellation? They will be closer, and their positions and alignment will form some (slightly) recognizable shapes. We can use shapes and distances between stars to detect which constellations stars belong to. If you know the constellations, you can easily see them by glancing at the night sky. But your brain already knows the forms of the constellations. A computer does not know them; therefore, we need to teach it. After learning the fundamentals, the computer just needs to repeat the math. And computers are particularly good at making calculations! Suppose you can detect a constellation in 1 s. In that case, a computer with a good algorithm could detect billions of constellations in less than a second.
Imagine we are looking at a shining star on a beautiful night and we see that it belongs to the Capricornus constellation. Now, imagine that we have the magic power to move stars—to push them in random directions. Suppose we want to move our star to the Sagittarius constellation. So, using our power, we move our star several times, until it stops closer to Sagittarius (Figure 3).
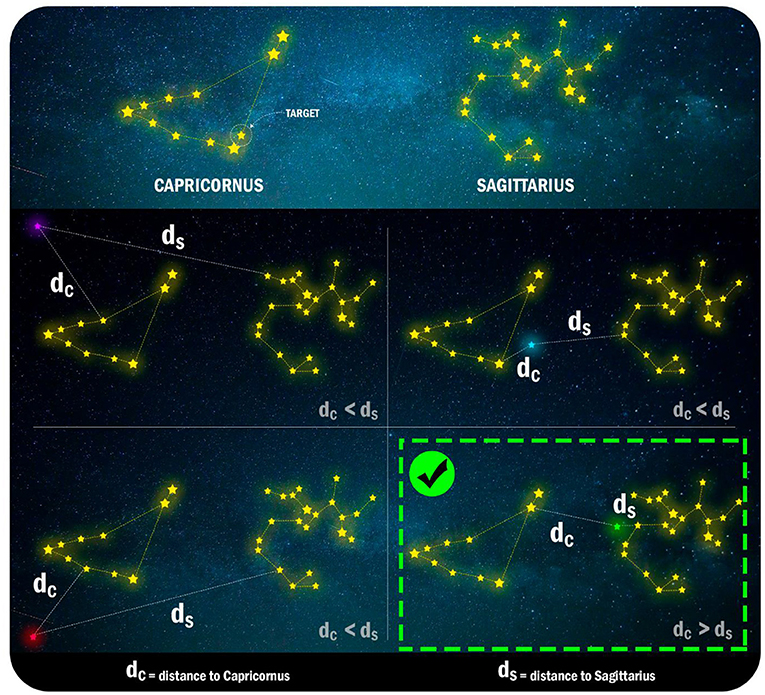
- Figure 3 - Yellow lines connect the stars that are in the same constellation.
- Each panel of the figure represents a use of our “magic power” to move stars. These movements correspond to mutations of an enzyme. After each movement, we calculate the distance between the closest stars from each constellation. Note that the purple, blue, and red stars stay closer to Capricornus, while only the green star moves closer to Sagittarius. Therefore, only the green mutation event changes the star’s constellation. When mutating enzymes, this would mean modifications in the structure of these molecules.
The same analogy can be made for computer simulations of enzyme mutations. Each star represents an enzyme used in biofuel production. The Capricornus constellation represents a set of enzymes that are inefficient for biofuel production. Sagittarius represents efficient enzymes. The “magic power” of moving stars represents the computer’s ability to simulate mutation. Mutations can occur naturally, but this process depends on many factors and can take millions of years. Genetic engineering allows scientists to quickly insert mutations into the structure of a molecule, allowing it to improve its activity (or decrease).
We simulate mutations by changing random parts of the enzyme. Then, we observe whether the mutated enzyme has a signature pattern closer to the known efficient or inefficient enzymes. If the mutation makes an enzyme’s signature pattern more similar to that of the efficient enzymes, we can assume that the mutant enzyme will have characteristics similar to the efficient ones. Then, scientists can make and test this mutation in the lab, to see if the mutated enzyme actually is more efficient at biofuel production.
Conclusion
Over the last several years, many studies have been done to improve enzymes for biofuel production. However, laboratory tests are expensive and take a lot of time. Using computer simulations, we can run millions of tests in seconds. Although they are not as accurate as laboratory tests, computer results can help scientists to figure out which laboratory tests are likely to give them positive results. Designing algorithms for biological purposes is not an extraordinarily complex task if you understand the biological problem well and know a programming language (we recommend Python).
Computers are one of humanity’s most amazing technological advances. They are responsible for a revolution in life sciences, helping scientists to improve biotechnological products like biofuels, bread, wine, cheese, yogurt, and many other foods, and to discover important new vaccines and drugs. To continue on this path, we need professionals with strong backgrounds in both computers and biology. Does that sound like you? If so, someday you can contribute to scientific discoveries that could change the world. So, come on, join us!
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001. Process numbers: 51/2013 - 23038.004007/2014-82 (CAPES) and 2016/17582-0 (FAPESP).
Glossary
Biofuel: ↑ Fuels made from organic matter, such as plants. First-generation biofuels are generally those most easily obtained, such as from sugarcane juice. Second-generation biofuels are those produced from organic matter, as those remaining from first-generation production.
Biomass: ↑ organic matter of plant or animal origin.
Fermentation: ↑ Chemical process of producing ethanol biofuel from sugars, generally made by enzymes from yeast or bacteria.
Saccharification: ↑ Process of extracting sugars (glucose molecules) from organic matter. This process can be done in several ways. For example, enzymes can be used to perform chemical reactions to break down the plant’s molecules, releasing glucose molecules.
Enzyme: ↑ A type of protein that speeds up chemical reactions. Also referred as molecules or macromolecules.
Genetic Engineering: ↑ Process of modifying the structure of a biological molecule (mutation) through laboratory experiments.
Algorithm: ↑ A step-by-step procedure used by a computer to solve a problem.
Signature Pattern: ↑ In structural bioinformatics, signature patterns are a set of characteristics obtained from computing analyses of some biomolecule. For example, counting the neighbor atoms number (the final list is the molecule signature).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the funding agencies: Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Original Source Article
↑Mariano, D. C.B., Santos, L. H., Machado, K. S., Werhli, A. V., de Lima, L. H. F., and de Melo-Minardi, R. C. 2019. A computational method to propose mutations in enzymes based on Structural Signature Variation (SSV). Int. J. Mol. Sci. 20:333. doi: 10.3390/ijms20020333
References
[1] ↑ Tester, J. W. 2005. Sustainable Energy. Cambridge, MA: MIT Press.
[2] ↑ Luterbacher, C., and Luterbacher, J. 2015. Break it down! How scientists are making fuel out of plants. Front. Young Minds. 3:10. doi: 10.3389/frym.2015.00010
[3] ↑ Santos, F. A., de Queiróz, J. H., Colodette, J. L., Fernandes, S. A., Guimarães, V. M., and Rezende, S. T. 2012. Potential of sugarcane straw for ethanol production. Quim. Nova 35:1004–10.
[4] ↑ Costa, L. S. C., Mariano, D. C. B., Rocha, R. E. O., Kraml, J., da Silveira, C. H., Liedl, K. R., et al. 2019. Molecular dynamics gives new insights into the glucose tolerance and inhibition mechanisms on β-glucosidases. Molecules 24:3215. doi: 10.3390/molecules24183215
[5] ↑ Mariano, D. C. B., Leite, C., Santos, L. H. S., Marins, L. F., Machado, K. S., Machado, K. S., et al. 2017. Characterization of glucose-tolerant β-glucosidases used in biofuel production under the bioinformatics perspective: a systematic review. Genet. Mol. Res. 16:1–19. doi: 10.4238/gmr16039740
[6] ↑ Mariano, D. C.B., Santos, L. H., Machado, K. S., Werhli, A. V., de Lima, L. H. F., and de Melo-Minardi, R. C. 2019. A computational method to propose mutations in enzymes based on Structural Signature Variation (SSV). Int. J. Mol. Sci. 20:333. doi: 10.3390/ijms20020333