Abstract
Did you know that your DNA can be read? We know that DNA is a storehouse of information that makes us who we are. But to truly understand information, we must first learn to read it. Genes are sections of DNA that, together, act as an instruction manual governing our cells, just like the codes that tell a computer what to do. For many years, we had no way to read these genetic codes. In the 1970s, the invention of a technology known as DNA sequencing made reading the DNA possible. DNA sequencing allowed scientists to read, understand, and compare genetic information, which was a major breakthrough in our understanding of biology. Today, sequencing technologies play an important role in everything from disease treatment to agriculture. In fact, DNA sequencing was crucial during the COVID-19 pandemic, helping us to study the coronavirus and rapidly develop vaccines.
The Discovery of ATGC
Deoxyribonucleic acid (DNA), which is the genetic material of every living organism, was discovered in 1869 by Swiss researcher Friedrich Miescher, who was originally trying to study white blood cells. Instead, he isolated a new molecule, which he called nuclein, from the nuclei of cells. The structure of the DNA molecule was identified by James Watson and Francis Crick in 1953, along with some major contributions by Rosalind Franklin [1]. The structure of DNA resembles a twisted ladder (Figure 1). The side rails of each step of the ladder are made up of a phosphate group (PO) and a pentose (five-carbon) sugar molecule, while the rungs are formed by four substances called nitrogenous bases, namely adenine (A), guanine (G), cytosine (C), and thymine (T). The pairings of these bases are explained by Chargaff’s rule, which states that A always pairs with T, and G always pairs with C. Together, a phosphate molecule, a pentose sugar, and a nitrogenous base make up what is called a nucleotide. The arrangement of the bases in DNA is very important because the sequence represents the genetic code, just like the proper arrangement of letters of the alphabet creates meaningful words. These “words” or blocks of information are the genes, and genes provide instructions to create various traits, from eye color to blood type. The collective term for all the genes present in the body is the genome. All members of a species have similar (but not identical) genomes. The human genome has properties that are unique to the human species, but small variations in the genes make every individual slightly different.
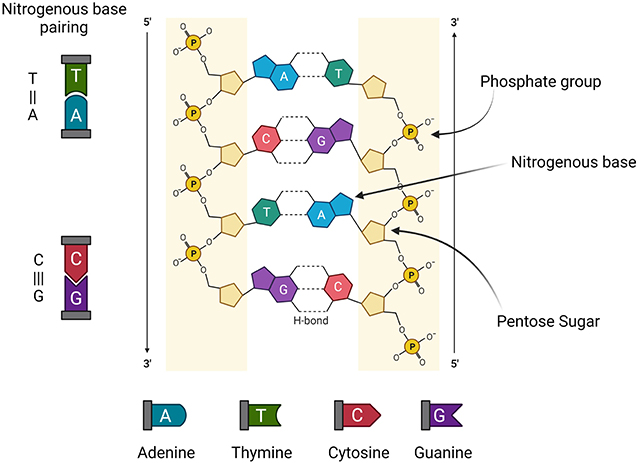
- Figure 1 - The double-helix, ladder-like structure of DNA determined by Watson and Crick.
- The sides of the ladder are made of pentose (five-carbon) sugars and phosphate groups. The steps of the ladder are the four nitrogenous bases: adenine (A), thymine (T), cytosine (C), and guanine (G). The bases pair to each other (A to T and C to G) through a type of molecular connection called hydrogen (H) bonding (Figure produced with https://biorender.com/).
First Attempt to Crack the Code: Sanger Sequencing
After the success of Watson and Crick’s findings, scientists had a clear understanding of the structure of a DNA molecule and its bases. Now came the question of determining the order, or sequence, in which the bases appeared along the length of the DNA. This process is called DNA Sequencing.
In 1975, Frederick Sanger made a huge breakthrough in DNA sequencing when he developed a method called chain termination [2]. In our cells, DNA replicates to make identical copies of itself using nucleotides (A, T, G, C) as building blocks. Sanger’s sequencing method was based on hijacking the natural process of DNA replication by adding chemically altered nucleotides, each with a radioactive label, in addition to the normal ones (Later the radioactive labels were replaced with fluorescent ones that are less dangerous). These modified nucleotides stopped the DNA replication at random points, whenever they were incorporated into the replicating DNA strand. Replication terminated in a random manner, creating several DNA fragments of different lengths, each ending with a modified, radioactively labeled (or color-coded) nucleotide.
Looking at all these fragments, Sanger could figure out the sequence of the original DNA strand by arranging them in the correct order, piecing together the fragments from smallest to largest, to visualize the complete DNA sequence. For example, the smallest fragment, only 1 nucleotide long, would be the modified version of A, T, G, or C. This told him the first base of the original DNA being sequenced (Figure 2). Sanger’s method of sequencing paved the way for large-scale DNA sequencing projects like the Human Genome Project.
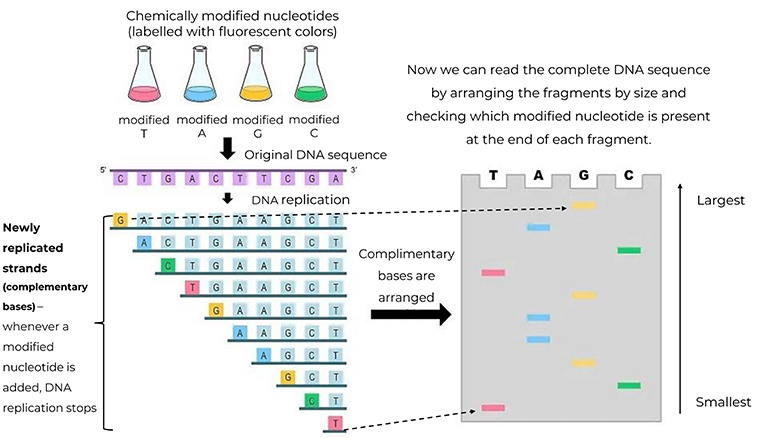
- Figure 2 - Chemically modified nucleotides (A, T, G, and C) labeled with fluorescent colors are added to the DNA replication mixture.
- Whenever a modified nucleotide is added to the DNA strand, replication stops. This produces complementary DNA fragments of varying lengths, which can be arranged to determine the original sequence of the DNA using the complementary base-pairing rule (A binds to T and G binds to C). This is a tedious process with many possibilities for error.
The Human Genome Project
The Human Genome Project (HGP) was a massive international project by scientists from all over the world, who began their research in 1990 and worked furiously for 13 long years to understand the human genome. They completed the sequencing of the massive human genome (containing 3.2 billion base pairs) in 2003—which was still ahead of schedule! The HGP was a landmark event. For the first time, scientists could read the entire genetic blueprint of humans, unlocking the doors to exciting new discoveries. Sequencing the complete human genome was a huge step in improving healthcare [3].
However, it is important to know that the DNA sequencing techniques used to achieve these results were time-consuming and inefficient. Despite the groundbreaking outcomes of the HGP, the birth of more advanced technologies, such as next-generation sequencing (NGS) has greatly improved the accuracy of DNA sequencing by minimizing errors. NGS has revolutionized the healthcare industry by lowering the cost of sequencing and the time needed for diagnosis. Today, the whole human genome can be sequenced in just 6 h!
What Is Next-Generation Sequencing?
In 2006, a company named Illumina announced a major advancement in DNA sequencing that could simultaneously sequence a large number of DNA strands at the same time. This high speed allowed the entire human genome to be sequenced in less than a day. This method differs from the earlier method of sequencing in that it does not depend upon chain termination. Instead, in this new method, whenever a new base is added to the replicating DNA strand, a corresponding fluorescent signal is emitted which is detected in real time (Figure 3).
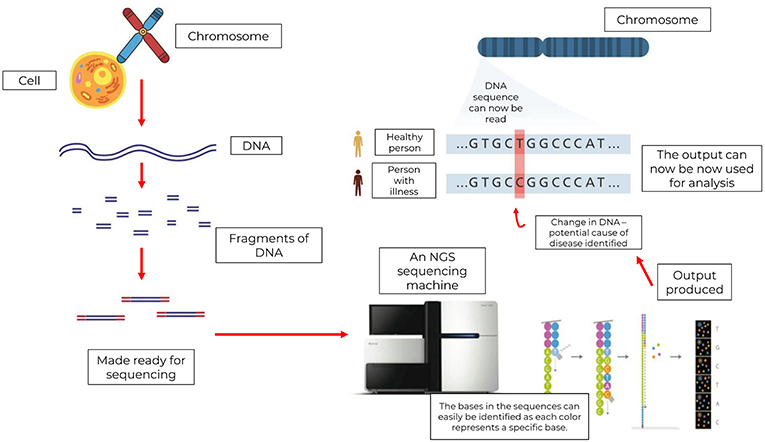
- Figure 3 - In NGS, many DNA strands are removed from cells and shredded into smaller fragments.
- These fragments are then made ready for sequencing by placing them on a plate called a flow cell, which contains the reaction mixture. The sequencer performs several chemical reactions and reads the sequences by identifying the fluorescent colors released from the flow cell whenever a new base is added. These signals can be interpreted by software to build a digital copy of the original DNA sequence. Scientists can now use this data to look for changes in genes that might cause illnesses like cancer.
How Is DNA Sequencing Used Today?
Cancer Treatment
Cancer has always challenged the medical world because each type of cancer is different and even the same cancer may act differently in different patients. Cancer can be caused by unique errors in the DNA that can be identified by DNA sequencing. Scientists are now using NGS to analyze the genomes of individual patients and to identify the DNA errors that are specific to each patient. This helps doctors design treatments customized to each individual. For example, two patients with the same type of cancer may respond differently to the same drug—a treatment that can cure one will fail on the other. Slight changes in the genomes of the two individuals can be detected using NGS and this can help doctors understand how to personalize therapy for individuals based on their genetic differences [4]. This type of customized treatment is called personalized medicine.
Evolution
Life on Earth started as single-celled organisms, which became increasingly complex. Animals are the most complex life forms on Earth. NGS has been used to compare the human genome with the genomes of other animals, which can tell us in fascinating detail about how closely or distantly we may be related to them. Additionally, small mutations (changes in the DNA) that exist in organisms of the same species are an important part of evolution because every mutation has the chance of permanently changing a part of that species’ genome. NGS allows scientists to study how mutations have accumulated throughout evolution.
Vaccines
From investigating the origin of the SARS-CoV2 virus to the rapid development of vaccines, sequencing has been a tremendous help during the COVID-19 pandemic. Vaccines teach the immune system to recognize threats and eradicate them. NGS helped scientists to sequence SARS-CoV2, which helped them learn more about the genetic features of the virus that threaten the immune system, such as the spike protein. NGS was also useful in tracking new viral variants—viruses evolve too!
Allergies
You have probably heard about gluten allergy or lactose intolerance. NGS helps scientists to identify whether an individual is likely to be allergic to gluten or intolerant to lactose, by analyzing the person’s genome. Certain DNA sequences might make a person more likely to develop allergies.
Helpful Microbes
Not all microbes living inside the body are harmful. Good microbes help us fight infections. The gut microbiota is a community of microbes that live in the human digestive tract and can be considered our “second genome” because they influence our immune systems and overall health [5]. The gut microbiota is not easy to study in labs, but NGS can be used to analyze the genomes of these organisms, to understand their functions. The American Gut Project1 was the world’s largest citizen science initiative that analyzed the microbes living inside the guts of roughly 11,000 people. It was made possible thanks to NGS.
Conclusion
In summary, DNA sequencing revolutionized life-science research and paved the way for more precise and efficient healthcare. NGS made this technique quicker and more affordable. In recent years, more advanced techniques of DNA sequencing like Oxford Nanopore were developed. These methods are even faster and more accurate! However, although we can sequence DNA, there still has been no method developed to sequence proteins. This is one of the most-awaited developments in biology. Hopefully one of you young readers will be able to achieve that grand feat!
Glossary
Nitrogenous Bases: ↑ Nitrogen-containing molecules that are important components of the DNA.
Nucleotide: ↑ Components of the DNA that contain a phosphate molecule, a sugar molecule, and a nitrogenous base.
Gene: ↑ A part of a DNA sequence that contains the code for making a protein.
Genome: ↑ The complete set of genes in a living organism.
DNA Sequencing: ↑ The process of determining the sequence of nitrogenous bases in a DNA fragment.
Human Genome Project: ↑ An international research effort that aimed at determining the sequence of all the genes in the human body.
Next-generation Sequencing: ↑ A modern and rapid method of DNA sequencing that can read a large volume of DNA sequences simultaneously.
Mutation: ↑ Any change or alteration in the DNA sequence that may or may not change the gene’s function.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnote
1. ↑See: https://www.facebook.com/AmericanGut.
References
[1] ↑ Watson, J., and Crick, F. 1953. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature 171:737–38. doi: 10.1038/171737a0
[2] ↑ Heather, J. M., and Chain, B. 2016. The sequence of sequencers: the history of sequencing DNA. Genomics 107:1–8. doi: 10.1016/j.ygeno.2015.11.003
[3] ↑ Green, E. D., Watson, J. D., and Collins, F. S. 2015. Human genome project: twenty-five years of big biology. Nature 526:29–31. doi: 10.1038/526029a
[4] ↑ Gupta, A. K., and Gupta, U. D. 2014. “Next generation sequencing and its applications,” in Animal Biotechnology (Elsevier). p. 345–67. doi: 10.1016/b978-0-12-416002-6.00019-5
[5] ↑ Cao, Y., Fanning, S., Pross, S., Jordan, K., and Srikumar, S. 2017. A review on the applications of next generation sequencing technologies as applied to food-related microbiome studies. Front. Microbiol. 8:1829. doi: 10.3389/fmicb.2017.01829

