Abstract
The evolution of bacteria is different from the evolution of other organisms. While we get our genetic information solely from our parents, bacteria can get part of theirs from other sources. In this article, we describe one of these chimeric bacteria, Azotobacter vinelandii, and explore how its dormant, seed-like form influenced its evolution.
Introduction
Earth’s first living cell appeared nearly four billion years ago. We call it LUCA, the last universal common ancestor. We do not know much about LUCA, but every plant, every animal, and every one of us humans evolved from LUCA. We know this thanks to studies done with special proteins contained in the cells of LUCA’s descendants. Proteins are indispensable for every living being, as they are the molecules that do the things that keep everything alive. Every biological process necessary for life, like respiration, digestion, or reproduction, involves proteins in some way. And while many of these processes and their corresponding proteins vary between organisms—for example, the respiratory proteins of fishes are very different from those of elephants—some proteins are so important that everyone has to produce them. One of these crucial functions for life is the mechanism used to produce proteins, which is carried out by a bunch of proteins coupled with other molecules and collectively called the ribosome. The ribosome is so crucial that it has not really changed much since LUCA was around. Tracking the small changes in the DNA that make the ribosome has allowed us to peek at the evolutionary history of living organisms and follow their relationships.
The Universal Tree of Life
Since all living organisms came from LUCA, and they all have ribosomes, it is possible to construct “family trees” that show us the relatedness of different organisms by tracking the small changes that have occurred in their ribosomes. In these family trees, which are called phylogenies by scientists, every species is located at the tips of the leaves, with the size of the branches between two tips indicating how similar the organisms at the tips are to each other. So, for example, the branches that separate cats, lions, and tigers are smaller than the branch that separates these animals from whales. Thus, in a mammals phylogeny all the felines form a tight group of branches that forks at the base from the whales’ branch. You can understand how a phylogeny is constructed using words, as describe in Figure 1A. If we grow the size of a phylogenetic tree by adding every known living being to it, we end up with the universal tree of life (UTL), in which we can see the relationships between all of the living beings [1] (Figure 1B).
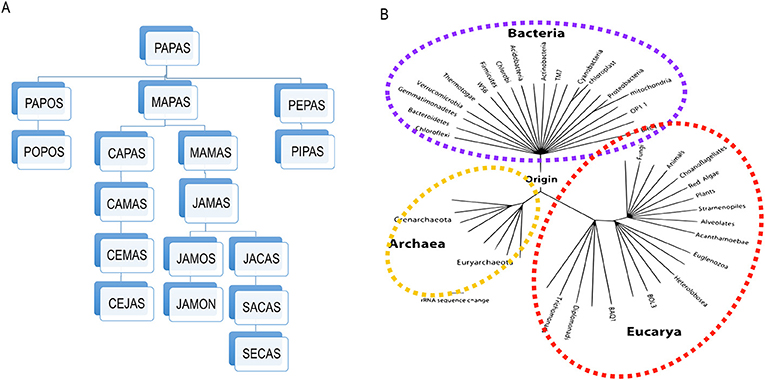
- Figure 1 - Building phylogenetic trees.
- (A) An example of how phylogenetic trees are constructed, using five-letters words. In this example, each change of a letter represents one unit of distance, so any two words that are just one letter/step apart in the tree are very similar, while those that are separated by more letters/steps (like “cejas” from “secas,” which are nine steps apart) are less related. You can see that the most recent common ancestor of any two words is where their paths converge (“mapas” in the case of “cejas” and “secas”). (B) A diagram of the tree of life built using the genetic sequence of the ribosome, for species belonging to the three domains of life: Bacteria, Archae (that are also unicellular microscopic organisms), and Eukarya (that include all animals and plants). Since all living organisms are included in this phylogenetic tree it is called the “universal tree of life” (this figure is a modification of a diagram from Pace in 2009).
In the example shown in Figure 1A we use five letter words, but you can imagine that, if you use larger words, you will be able to measure greater distances. The full length of the gene for the bacterial ribosome is around 1,500 “letters,” so the calculation of the relationships between organisms using the ribosome is very sensitive and thus all organisms can be included in the UTL.
The path formed from the bottom of the tree to the leaf tips represents the evolutionary path that led to the species at that tip. The members of that species, having followed that specific path, lost the capacity to mate and have offspring with members of other species that followed different evolutionary paths.
Bacteria Not Always Inherit Their Genes From Their Parents
Every species inherits its DNA, or genetic information, directly from its ancestors. Those ancestors got their DNA from the generation that came before them, and so on, and so on, tracing the evolutionary path backward until the tree is traced all the way back to LUCA. Because the genetic information of each species is unique to that species, it can only be shared with the members of that same species, flowing from the parents to their offspring. That is, unless we are talking about bacteria. Bacterial species have the ability to share parts of their genetic information with other bacterial species that have not followed the same evolutionary paths; bacteria are able to inherit DNA from other bacteria that are not in any way related to them. This movement of genetic information is called horizontal gene transfer (HGT), since the transfer of genetic information occurs horizontally, between bacteria of the same generation, instead of vertically, between parents and offspring.
Another interesting characteristic of bacteria is that some of them can develop dormant forms that are similar to the seeds of plants. These dormant forms can exist for a long time without reproducing (Figure 2). The dormant forms are like an ancient reservoir of genetic information that, in theory, could be harnessed by other organisms [2]. The genetic information of the dormant forms is preserved for many years without change and can be incorporated into other bacteria by HGT.
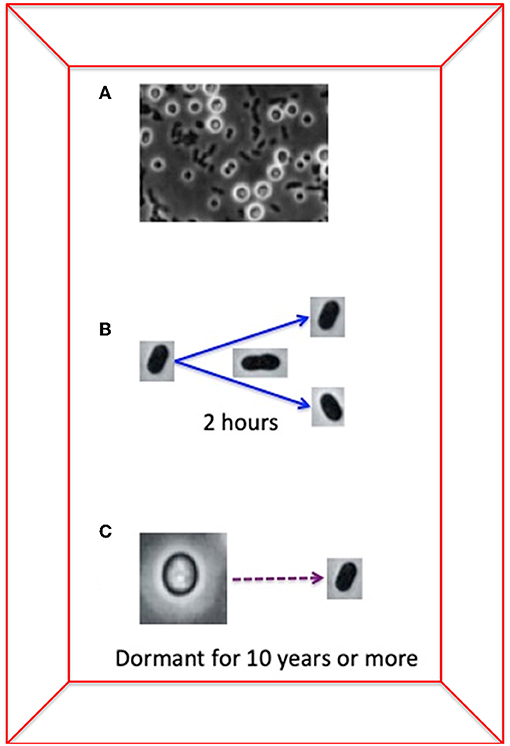
- Figure 2 - The chimeric bacterium Azotobacter vinelandii has two forms of reproduction: its reproduction by cell division takes 2 h, while dormant, seed-like structures can be alive without reproducing for more than 10 years.
- (A) A picture (taken using a light microscope) of bacterial cells and the dormant, seed-like form. You can compare the size and form of the two types of cells: the seed-like form is much larger, while normal bacterial cells are small and dark. (B) Azotobacter vinelandii takes 2 h to reproduce by cell division. (C) The dormant, seed-like form of Azotobacter can persist in dry conditions for 10 years or more. So, Azotobacter vinelandii “seeds” remain dormant for 44,000 times longer than it takes the cells to reproduce by cell division! It is possible that the genetic information in dormant forms results in a reservoir of genes that can be incorporated from distantly related bacteria by HGT.
Are Bacteria Able to Form Chimeras?
To have a clearer picture of how dormant forms of bacteria could affect bacterial evolution, let us imagine that dinosaurs could form eggs that could remain in the environment for tens of millions of years. In that case, once in a while we would bump into a baby dinosaur, which would contain genes that are not present in any other organism that is living today, and would coexist with modern animals. Let us imagine further that these newly born dinosaurs could mate with other animals. Then it would be possible to create chimeric animals, which are animals that contain genetic information, and therefore the physical parts, of different species. In this scenario, a chimeric animal with parts of birds, dinosaurs, and mammals could exist!
We imagined that the ability of bacteria to inherit DNA by HGT and their capacity to stay dormant for long periods, will permit that bacterial chimeras are formed, just as in the example of the baby dinosaur. This is the question that driven our research [2].
In the Search of a Bacterial Chimera: The Case of Azotobacter vinelandii
We were interested to find out whether the dormant forms of bacteria could create chimeric bacteria, using their ability to transfer genetic information through HGT. To see if this could happen, we looked at the genome of Azotobacter vinelandii, which is a bacterium that is able to form dormant forms called chists (Figure 2), and is very closely related to another bacterium called Pseudomonas aeruginosa [3]. It is a little bit of shock that these organisms are related, frankly, because these two species of bacteria are very different from each other! We investigated this interesting fact and found that these organisms are related on the tree of life because nearly half of Azotobacter vinelandii’s genetic information, including some of the instructions for making ribosomes, is closely related to that of Pseudomonas [4], while the rest of the information is used to make the proteins that give Azotobacter its own special characteristics. As if that was not enough, we also found that the genetic information that did not come from Pseudomonas was inherited from different unrelated bacterial species [2]. In other words, the core of the genetic information of Azotobacter vinelandii came from a shared ancestor of Azotobacter and Pseudomonas, but during its particular evolutionary path, Azotobacter acquired additional genetic information that made it very different from its “cousin” Pseudomonas, or to any other organism really, because Azotobacter is a chimera.
How Common Are Bacterial Chimeras?
While it is difficult to know if chimeric organisms are common, the existence of Azotobacter is strong evidence that chimeras might not be that rare. There are at least two other well-documented cases of bacterial species that seem to have a core derived from a bacterial group called Firmicutes and other pieces of genetic information obtained from other organisms. These chimeras are bacteria called Thematogales, organisms that acquired genes that allowed them to live at high temperatures, and Fusobacterium nucleatum, a bacterium found in dental plaque that acquired genes from neighboring bacteria. Neither Thermatogales nor Fusobacterium nucleatum form dormant seed-like structures, but the information that they acquired by HGT might have come from an ancient bacteria that did have the capacity to make dormant forms. We still do not know, but maybe someday we will.
Glossary
Proteins: ↑ Large, complex molecules that play many critical roles in the body. Proteins do most of the work in cells and are required for the structure, function, and regulation of the body’s tissues and organs.
Ribosomes: ↑ Ribosomes are complex structures formed of proteins and other molecules. These molecular machines are present in every cell and are the site where new proteins are produced.
Genetic Information: ↑ The DNA in every organism, in which the information required to make proteins and other cellular components is stored. DNA is passed from one generation to the next through inheritance.
Horizontal Gene Transfer (HGT): ↑ The process of inheriting genetic information from an unrelated species. This mechanism of inheritance is not common, since organisms usually inherit their genetic information from their parents that are members of the same species as their offspring.
Chimera: ↑ Any mythical animal formed from parts of various animals. This term is used in biology to describe an organism containing a mixture of genetically different tissues. In this article, we use this term to describe bacteria with genetic information inherited from different organisms, some of which are not their ancestors.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I deeply acknowledge the enthusiastic and fruitful discussions with Karla Galaviz, Adrián González-Casanova, Inés González-Casanova, and Ximena Martínez de la Escalera.
Original Source Article
↑González-Casanova, A., Aguirre-von-Wobeser, E., Espín, G., Servín-González, L., Kurt, N., Spanò, D., et al. 2014. Strong seed-bank effects in bacterial evolution. J. Theor. Biol. 356:62–70. doi: 10.1016/j.jtbi.2014.04.009
References
1. ↑ Pace, N. 2009. Mapping the tree of life: progress and prospects. Microbiol. Mol. Biol. Rev. 73:565–76. doi: 10.1128/MMBR.00033-09
2. ↑ González-Casanova, A., Aguirre-von-Wobeser, E., Espín, G., Servín-González, L., Kurt, N., Spanò, D., et al. 2014. Strong seed-bank effects in bacterial evolution. J. Theor. Biol. 356:62–70. doi: 10.1016/j.jtbi.2014.04.009
3. ↑ Rediers, H., Vanderleyden, J., and De Mot, R. 2004. Azotobacter vinelandii: a Pseudomonas in disguise? Microbiology 150:1117–9. doi: 10.1099/mic.0.27096-0
4. ↑ Martínez-Carranza, E., Ponce-Soto, G. Y., Servín-González, L., Alcaraz, L. D., and Soberón-Chávez, G. 2019. The evolution of bacteria seen through their essential genes: the case of Pseudomonas aeruginosa and Azotobacter vinelandii. Microbiology 165:976–84. doi: 10.1099/mic.0.000833