Abstract
Did you know that the earth beneath your feet is teeming with life? Imagine yourself standing outside. If you started digging, at some point beneath the soil you would eventually hit hard rock—that is the bedrock forming the Earth’s crust. Even this seemingly solid material has cracks and pores that contain groundwater (water that is held in the ground), and where there is water, there can be life. Tiny, single-celled creatures called microbes survive and thrive in many environments on Earth that are inhospitable for all other life forms. This is also the case with deep bedrock, where only microbial life is possible. As sunlight and plant products are not available in this environment, microbes have to use chemical compounds for their food and energy sources. In this study, we investigated the preferred food for microbes living in the bedrock. We discovered that hydrogen is an important energy source for deep microbes, and that the microbial communities are able to change so that they can survive under changing environmental conditions.
Introduction
Life on the surface of the Earth is dependent on the energy of the Sun, which allows green plants to produce sugars and oxygen. However, a large amount of Earth’s total biomass (living material) is located in the deep subsurface—this means inside the Earth’s rocky crust. Bedrock is not a big lump of solid rock; instead, it contains cracks, fractures, and openings that can be filled with water. Recent studies have shown that some of these fractures can contain fluids that are more than one billion years old [1]. This leaves us with a very interesting thought: maybe microbes have been living in these environments as long as the water has been there! If so, we can ask how these microbial communities survive in these habitats.
Microbial Food—Energy Sources in the Deep, Dark Bedrock Environment
Many subsurface (underground) environments are completely cutoff from the surface world, which means that energy and food for microbes dwelling in the depths must come from the surrounding rocky environment. Because of this, deep subsurface environments are usually oligotrophic, meaning that they lack nutrients, and if these environments do have nutrients, the quantities are so low that we cannot even detect them. However, microbes are so tiny that they only need very small amount of nutrients, so they are equipped to survive in these environments.
Carbon is the building material of cells, and microbes can use different types of carbon materials for food and energy sources. Some microbes use carbon dioxide (CO2) gas, as plants do, but others prefer organic carbon sources, such as sugars, fatty acids, or amino acids, which originate from living material. Respiration is a process in which chemical energy is released into use of the organism. For example, humans breathe in oxygen and use it to break down sugar (chemical energy), which then makes our muscles move (biological energy). However, in the deep subsurface, microbes need to use other respiration mechanisms, since there is no oxygen around. Respiration that occurs in the absence of oxygen is called anaerobic respiration. A common anaerobic respiration mechanism is called sulfate reduction.
Hydrogen and CO2 are frequently found in deep subsurface environments, because they are formed in the Earth’s crust or mantle. Microbes that can use hydrogen and CO2 for energy are known to exist in the deep subsurface. These microbes are called autotrophic chemolithotrophs (or chemolithoautotrophs), which means that they only use CO2 as a carbon source (autotroph) and other molecules that do not contain carbon for their energy (chemolithotroph, lithotroph is derived from some Greek words that mean “rock-eater”).
We studied whether carbon from other living things, such as plants or animals (called organic carbon), or carbon from non-living sources such as minerals (called inorganic carbon) is the favored carbon source for the bacteria that live deep in the bedrock groundwater. Microbes from this deep groundwater were fed either only inorganic carbon (in the form of CO2) or both inorganic and organic carbon (in the form of a molecule called acetate) (Figure 1). We also tested the microbial communities for their ability to produce energy via sulfate reduction.
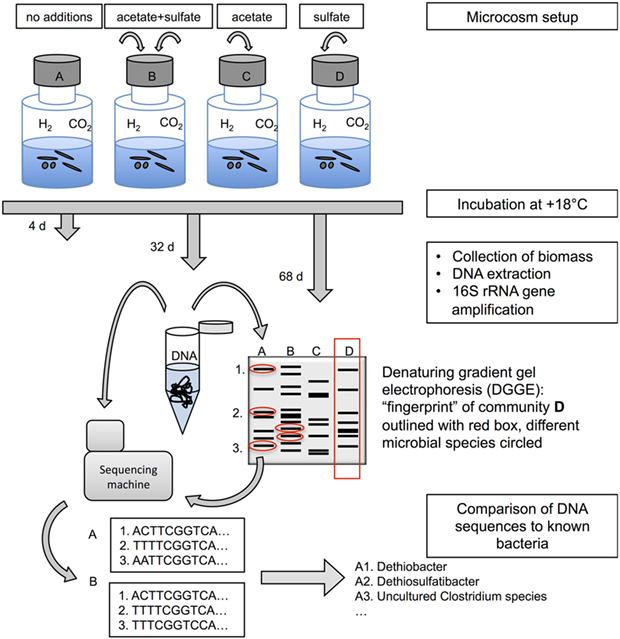
- Figure 1 - A schematic representation of the design of the study and different methods used.
- Topmost part of this figure shows the microcosm setup with glass bottles where we put the groundwater and gave microbes different meals: (A) only inorganic carbon source (CO2) and energy source (H2) in gas phase, both naturally found in deep subsurface gases; (B) acetate as an organic carbon source and sulfate as an energy source; (C) acetate as an organic carbon source; and (D) sulfate as an energy source. The middle and lower parts of this figure describe how DNA extracted from the microbes in the microcosms is subsequently being analyzed with fingerprinting and sequencing methods.
Sampling the Deep Bedrock and Setting Up the Experiment
In this study, we obtained groundwater samples from the deep bedrock environment (a depth of 967 m) by pumping fluids from the Outokumpu scientific deep drill hole, located in Eastern Finland. Sampling was done by pumping the fluid in plastic tubing from the subsurface to the field laboratory, where we had a portable anaerobic (oxygen-free) chamber, so that we could work with the samples in an oxygen-free environment. This is important, because oxygen is toxic to some of the microbes living in the deep groundwater. We split the groundwater up into separate bottles, as shown in Figure 1. Each bottle was treated a little differently—they were given different carbon and energy sources. Each bottle in this kind of experimental setup is called a microcosm, and we used these microcosms as a way to study microbes that need different carbon and energy sources. The gas above the liquid in each microcosm contained a mixture of hydrogen and carbon dioxide. An organic carbon source called acetate was added to some microcosms, either with or without sulfate. Microcosms with an inorganic carbon source (CO2) and sulfate were also prepared. These microcosms were incubated at the same temperature (+18°C) that was present in the drill hole, for 4, 32, and 68 days. Samples were collected at each of these time points, and DNA was extracted to see what kind of microbes were growing.
Microbes Were Studied by Looking at Their DNA
Microbial communities can be characterized using certain tests that look at the DNA molecules of the microbes. DNA, as you might already know, is the genetic material of the cell. It contains instructions, in the form of genes, which provide the blueprint for creating the organism. Studying specific genes is a good way for scientists to learn about microbial communities. For example, we can look at a gene called the 16S rRNA gene. The 16S rRNA gene tells us about the relationships of microbes, and that is why it is called a “phylogenetic” marker gene. “Phylogenetic” is derived from Greek words meaning the origin of a tribe or a family. There were other marker genes used in this study, too, including genes coding for special proteins called enzymes—specifically enzymes that are important in using sulfur for energy.
By studying the DNA of these microbes using several different methods, which will be described below, we were able to study three things: how many microbes were present in the samples taken from the groundwater, how those microbes grew over time in each microcosm, and how those microbes were related to each other and to other microbes that we know about.
How Many Bacteria Were There in the Deep Groundwater Samples?
With a laboratory technique called quantitative polymerase chain reaction, we calculated how many copies of the bacterial genes were in each microcosm, by comparing the results to standards containing a known amount of gene. This experiment gave us data about the numbers of bacteria present in the groundwater samples and also allowed us to see how the microbes did under the different experimental conditions tested in each microcosm.
We detected an increase in the copy numbers of the phylogenetic marker gene (the 16S rRNA gene) during the course of the experiment (Figure 2). The copy numbers were around 500/ml in the beginning of the experiment and increased to several tens of thousands to as high as one million per milliliter during the experiment. This means that microbial cells in the microcosms were multiplying during the incubation period. The copy numbers of another gene we looked at, the sulfate reduction marker gene, also increased in the microcosms that had sulfate and acetate added to them. We also saw an increase in bacterial numbers in the microcosms that did not receive anything. In this case, we think that some of the original microbes died during the experiment and others used the material from the dying cells as their food and as the building blocks for new cells.
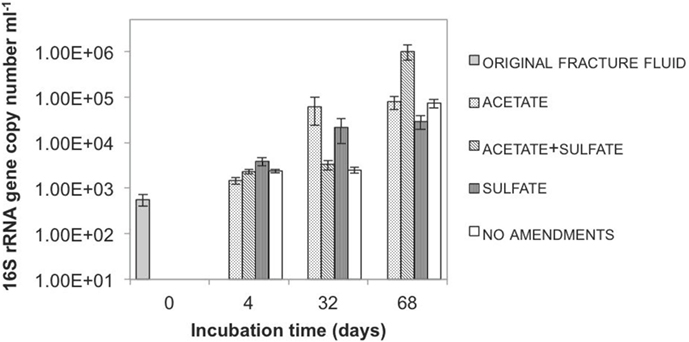
- Figure 2 - Quantity of microbes in different microcosms during the incubation.
- Each column represents the quantity (y-axis) of bacteria in a specific microcosm experiment at a given time point (x-axis). Figure modified from Ref. [2].
The DNA “Fingerprints” of the Microbial Microcosms Can Tell Us How They are Related to Each Other
The DNA code of the 16S rRNA gene varies slightly from one microbial species to another, and because of this, pieces of DNA from different microbes can be separated from each other with a method called denaturing gradient gel electrophoresis (Figure 1). DNA strands, which always have a negative charge, travel in the gel toward a positive charge at the bottom of the gel. The small differences in the DNA sequences between different microbes will make them travel through the gel at slightly different speeds, so they will separate from each other, forming different bands in the gel. Each band represents a specific bacterial species that is present in the microcosm. This “fingerprint” can then be visualized under ultraviolet light.
Microbial Communities are Constantly Changing
We used DNA fingerprint method to study how the microbial communities changed in time in the microcosms. We noticed that the number of bands in the fingerprint of each microcosm decreased during the incubation period. This means that the number of different kinds of bacteria decreased during the experiment. The fingerprint patterns of microbial communities divided the microcosm samples into two groups: one group included the intrinsic fracture fluid sample and the microcosms incubated for 4 and 32 days and the other consisted of the microcosms after 68 days of incubation. This means that the incubation time had a more significant effect on the bacterial community than the different conditions present in each microcosm.
There are Lots of Different Microbes Present in the Bedrock!
We can “read” the DNA code of bacteria with a method called sequencing. Comparing the DNA codes, we can see how the organisms from the deep groundwater samples are related to each other and how they are related to other microbes in that scientists already know about. We used this technique to understand what kinds of microbes were present and how different microbial communities were in the microcosms.
The bacterial communities in the microcosms with added carbon and energy sources were more diverse in the first two time points compared with the natural groundwater microbial community and the microcosm experiment that did not receive any additions (Figure 3). However, in the last time point, the most diverse bacterial community was found from the microcosm that had not received any nutrients. The most frequently detected microbial species in the microcosms during the first 4 weeks were certain types of microbes that use hydrogen to obtain energy. The most significant change in the bacterial communities in this study was observed after 68 days. After 68 days of incubation, the hydrogen-using microbes had disappeared, and the most frequent species was a different one that did not use hydrogen. This might have occurred because the hydrogen-using microorganisms started to die off, and those dead microbes served as food for microbes that typically eat decomposing organic matter, such as dead microbial cell material. Many of the bacteria that we detected were closely related to microbes found from other subsurface and groundwater environments.
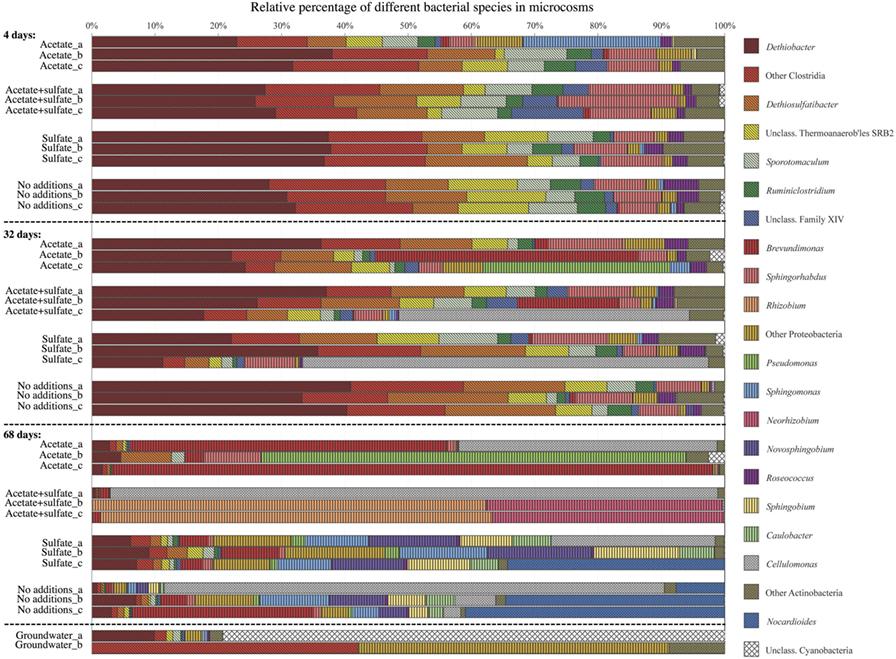
- Figure 3 - Microbial community structure in four different treatments in three time points, detected from triplicate samples (a–c).
- Each bar represents the bacterial community in the microcosm, and within each bar, different types of bacteria are labeled with different colors. For comparison, the groundwater microbial community without any additions or incubation is depicted in the bottom (duplicate samples a and b). Figure modified from Ref. [2].
Rock-Derived Hydrogen—A Snack for the Deep Microbes
From our experiments, we could see that hydrogen addition was the most important factor in molding the microbial communities. Hydrogen-using bacteria (called Dethiobacter) dominated our microcosms. Our previous study showed that these microbes are playing a big role in formation of the microbial community in deep bedrock [3]. These organisms were also found by another study describing microbial communities enriched from serpentinizing groundwater [4]. Serpentinization is a reaction where rock and water interact, and the minerals in the rock produce a significant amount of hydrogen. So, serpentinization reactions can provide a source of energy and raw materials to support chemolithotrophic microbial communities. We have not actually seen that serpentinization is going on in the Outokumpu bedrock, but there are some rocks in the bedrock that tell us that serpentinization has occurred in the past, meaning that it may have left behind a suitable environment for these hydrogen-using microbes.
What Did We Learn About Microbes in Deep Groundwater?
We concluded that organic carbon and hydrogen are useful for microbes in deep subsurface environments, but although these environments are thought to be very stable, there are probably cyclic changes, meaning that different microbial groups dominate at different times, providing material to each other. When viruses attack or food is used up and one type of microbe begins to die off, other microbial groups will survive and thrive.
Glossary
Deep Subsurface: ↑ Is a part of Earth that extends from few tens of meters to several kilometers depth into the Earth’s crust.
Autotrophic: ↑ Microbes use CO2 as a carbon source.
Chemolithoautotroph: ↑ Is an organism that obtains its metabolic energy from the oxidation of reduced inorganic compounds and its carbon for biosynthesis from CO2.
Microcosm: ↑ Is an artificial ecosystem used to study and simulate natural ecosystem in a controlled laboratory conditions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Original Source Article
↑ Purkamo, L., Bomberg, M., Nyyssönen, M., Ahonen, L., Kukkonen, I., and Itävaara, M. (2017). Response of deep subsurface microbial community to different carbon sources and electron acceptors during ∼2 months incubation in microcosms. Front. Microbiol. 8:232. doi:10.3389/fmicb.2017.00232
References
[1] ↑ Holland, G., Sherwood Lollar, B., Li, L., Lacrampe-Couloume, G., Slater, G. F., and Ballentine, C. J. 2013. Deep fracture fluids isolated in the crust since the Precambrian era. Nature 497:357–60. doi:10.1038/nature12127
[2] ↑ Purkamo, L., Bomberg, M., Nyyssönen, M., Ahonen, L., Kukkonen, I., and Itävaara, M. 2017. Response of deep subsurface microbial community to different carbon sources and electron acceptors during ∼2 months incubation in microcosms. Front. Microbiol. 8:232. doi:10.3389/fmicb.2017.00232
[3] ↑ Purkamo, L., Bomberg, M., Kietäväinen, R., Salavirta, H., Nyyssönen, M., Nuppunen-Puputti, M., et al. 2016. Microbial co-occurrence patterns in deep Precambrian bedrock fracture fluids. Biogeosciences 13:3091–108. doi:10.5194/bg-13-3091-2016
[4] ↑ Crespo-Medina, M., Twing, K. I., Kubo, M. D. Y., Hoehler, T. M., Cardace, D., McCollom, T., et al. 2014. Insights into environmental controls on microbial communities in a continental serpentinite aquifer using a microcosm-based approach. Front. Microbiol. 5:604. doi:10.3389/fmicb.2014.00604

